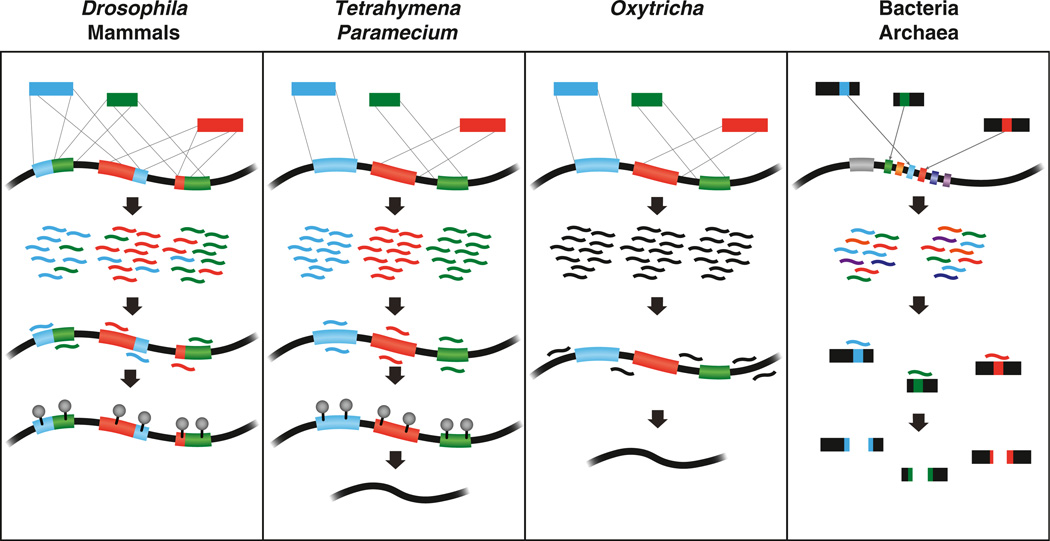
Figure 3. Small RNA-Mediated Genome Defense.
Foreign transposable and repetitive sequences, represented by blue, green, and red segments, are present in the genomes of mammals, insects, and ciliates. Mammals and Drosophila utilize piRNAs to silence active transposons, either by DNA methylation or by repressive histone modifications (gray circles). In Tetrahymena and Paramecium, scnRNAs recognize IES regions, which include mobile genetic elements, and guide their genomic excision by marking them with repressive histone modifications. Oxytricha eliminates IES sequences using an orthogonal mechanism; piRNAs correspond to the retained sequences (black). Chromatin modifications that guide this process have not yet been identified. Bacteria and archaea incorporate sequences from foreign pathogens or plasmids (colored segments) into CRISPR loci. Expression of crRNAs from these loci results in cleavage of infecting bacteriophages and plasmid DNA.