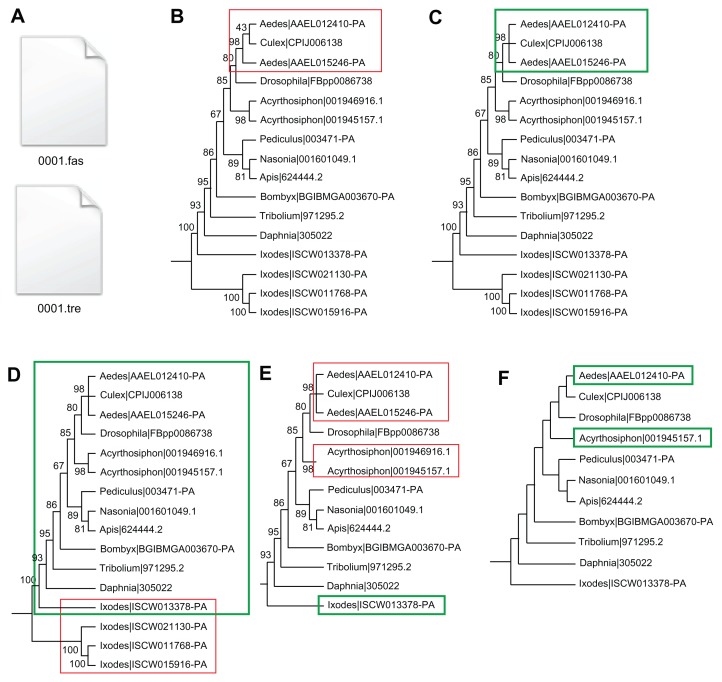
Figure 1.
Illustration of the PhyloTreePruner tree-pruning algorithm. (A) PhyloTreePruner reads the single-gene tree and corresponding alignment file. (B) Nodes in the single-gene tree with support values below the user-defined threshold are identified (red box) and (C) collapsed into polytomies (green box). (D) PhyloTreePruner identifies the maximally inclusive subtree in which all taxa are represented by exactly one sequence, or, if there is more than one sequence from a taxon, these sequences form a monophyletic clade or are part of the same polytomy (green box). In this example, PhyloTreePruner identifies a potential paralogy issue with the Ixodes sequences (red box). This example shows the necessity of correct single-gene tree rooting. (E) PhyloTreePruner deletes sequences inferred to be paralogs from the tree and the corresponding sequence alignment file (red boxes). (F) In cases where more than one sequence remains from the same taxon, PhyloTreePruner selects the longest sequence and deletes all others (green boxes). This step can be skipped if preferred and another method (eg, SCaFoS) can be used to select the best sequence for each taxon.