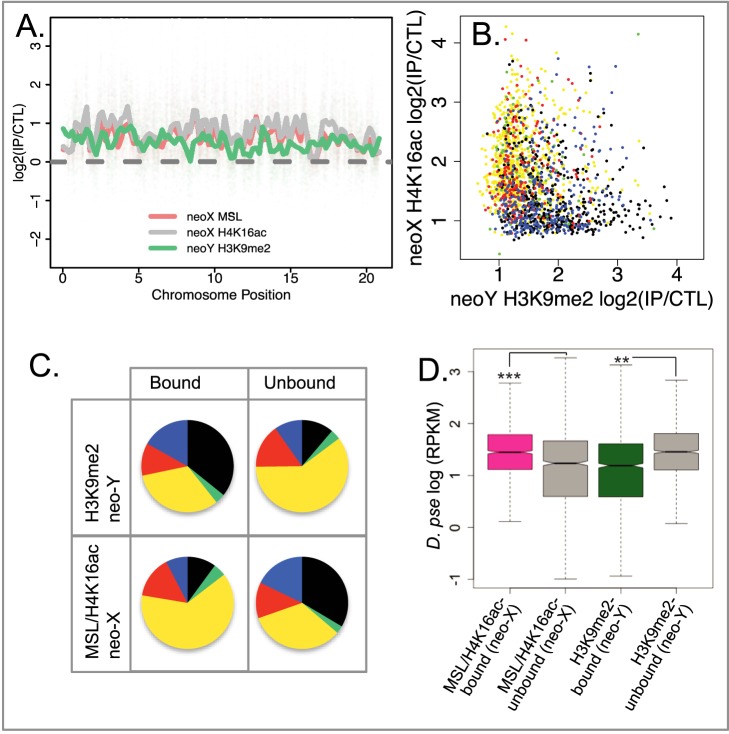
Figure 5. Heterochromatin formation and dosage compensation.
(A) Sliding window enrichment profile of H3K9me2-enrichment along neo-Y genes, and H4K16ac and MSL-binding along their neo-X homologs. (B) H4K16ac-enrichment of neo-X genes versus H3K9me2-enrichment at their neo-Y homologs. Genes are color coded according to their chromatin state in D. melanogaster [43], with yellow and red corresponding to genes located in active chromatin, and black, green, and blue corresponding to genes located in repressive chromatin. (C) MSL/H4K16ac-bound/unbound neo-X genes and H3K9me2-bound/unbound neo-Y genes versus principle chromatin types in D. melanogaster. The color-coded chromatin types of D. miranda bound/unbound genes were inferred from the chromatin type definition of their D. melanogaster orthologs (from [43]). Genes within “yellow” chromatin are more likely to be targeted by the dosage compensation complex on the neo-X. Genes within “black” chromatin are more likely to be silenced by H3K9me2 on the neo-Y. Genes within active “red” chromatin show no significant difference regarding their dosage compensation states on the neo-X, which is consistent with the lack of H3K36me3 chromatin mark in red chromatin [43], and the dosage compensation complex targeting genes with such a mark. (D) Expression levels of genes in D. pseudoobscura whose homologs in D. miranda are bound/unbound by MSL/H4K16ac on the neo-X or bound/unbound by H3K9me2 on the neo-Y; D. pseudoobscura expression levels can be used as a proxy for ancestral expression of neo-sex linked genes.