Summary
RAS proteins directly activate PI3-kinases. Mice bearing a germline mutation in the RAS binding domain of the p110α subunit of PI3-kinse are resistant to the development of RAS-driven tumors. However, it is unknown whether interaction of RAS with PI3-kinase is required in established tumors. The need for RAS interaction with p110α in the maintenance of mutant Kras-driven lung tumors was explored using an inducible mouse model. In established tumors, removal of the ability of p110α to interact with RAS causes long-term tumor stasis and partial regression. This is a tumor cell-autonomous effect, which is improved significantly by combination with MEK inhibition. Total removal of p110α expression or activity has comparable effects, albeit with greater toxicities.
Highlights
-
•
Inducible disruption of PI3-kinase p110α interaction with RAS was modeled in mice
-
•
Blocking RAS/p110α interaction in established lung tumors causes partial regression
-
•
Blocking RAS/p110α binding has effects similar to p110α deletion on tumor regression
-
•
Coordinate MEK inhibition is required for major regression of Kras mutant lung cancer
Significance
Blocking the interaction of PI3-kinase p110α with RAS as well as total loss of p110α or inhibition of its lipid kinase activity are shown to be beneficial in the treatment of established Kras mutant lung cancer. Because preventing RAS binding to p110α has similar effects on Kras mutant tumor regression to total removal of p110α, the interaction between RAS and p110α has central importance in maintaining PI3-kinase signaling in RAS-driven cancers. Blocking this could provide a less toxic approach to targeting these tumors than total PI3-kinase enzymatic inhibition. Furthermore, simultaneous inhibition of another major RAS effector pathway, RAF/MEK/ERK, together with blockade of either p110α or all type I PI3-kinases, causes major tumor regression.
Introduction
Activating point mutations in the genes encoding the RAS subfamily of small GTP binding proteins drive the formation of a large proportion of human tumors. RAS proteins control cell growth through several direct effector enzyme families, the best studied of which are RAF kinases, type I phosphoinositide (PI)3-kinases, and RAL-guanine nucleotide exchange factors (RAL-GEFs) (Downward, 2003; Pylayeva-Gupta et al., 2011). Of these, BRAF and PIK3CA, the genes encoding BRAF and the p110α PI3-kinase catalytic subunit, respectively, are frequently activated by somatic mutation in human cancer (Weir et al., 2004), with overall mutation frequencies around 10% for each. PI3-kinase activity is further implicated in carcinogenesis by the frequent inactivation of the inositol lipid 3′phosphatase PTEN (Cully et al., 2006).
The importance of the interaction of mutant RAS with endogenous p110α in tumor development was established by the findings that mice with mutations in the RAS binding domain (RBD) of p110α were highly resistant to mutant Kras-induced lung cancer formation and mutant Hras-induced skin cancer formation (Gupta et al., 2007). However, it has been speculated that the signaling pathway requirements for tumor cells to survive (tumor maintenance) may be considerably relaxed relative to the signaling pathway activity needed to convert a normal cell into a tumor cell (tumor development) (Lim and Counter, 2005). As tumor maintenance is more relevant than tumor initiation to the treatment of preexisting human cancers, the question of whether blocking the interaction of RAS with p110α will affect the maintenance of an existing RAS mutant tumor is of considerable importance.
Inhibition of PI3-kinase activity has been reported to have a significant impact on RAS-induced tumors in some settings, especially when combined with inhibition of the RAF/mitogen-activated protein kinase extracellular signal-regulated kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway (Engelman et al., 2008; Halilovic et al., 2010; She et al., 2010; Sos et al., 2009). However, it has been unclear whether PI3-kinase signaling is more important for RAS mutant tumors compared to tumors with other oncogenic drivers or to normal cells. Also, where such PI3-kinase dependency exists, it is not known whether this is because of acute RAS-induced activation of PI3-kinase through direct interaction with the RBD of p110 or more indirect and long-term mechanisms, such as transcriptional upregulation of ligands of growth factor receptor tyrosine kinases.
Here, we have set out to address the issue of the importance, or otherwise, of the direct interaction of RAS with the RBD of PI3-kinase p110α in the maintenance of RAS-induced tumors.
Results
Removal of RAS Interaction with PI3-Kinase p110α in Early-Stage Tumors Reduces Tumor Burden
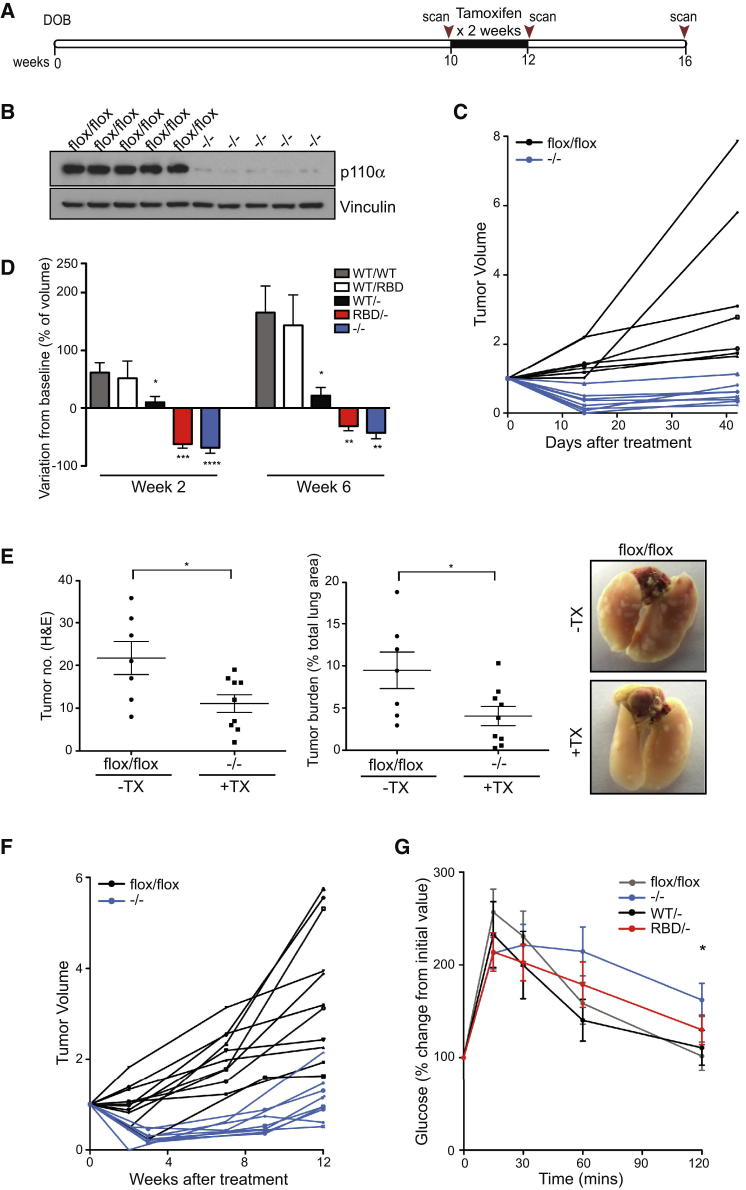
In order to investigate the role played by the direct interaction of p110α with RAS in tumor maintenance, we used a previously generated mouse model in which the interaction of p110α with RAS was disrupted by the introduction of two point mutations, T208D and K227A, into the endogenous Pik3ca gene (Pik3caRBD) (Gupta et al., 2007). Wild-type (Pik3caWT) and Pik3caRBD mice were bred with mice containing a floxed Pik3ca allele (Zhao et al., 2006) and a mouse carrying a conditional Cre recombinase (Cre-ERT2) allele targeted to the ubiquitously expressed Rosa26 locus. Finally, they were bred with KrasLA2 mice (Johnson et al., 2001) so that they spontaneously developed lung adenocarcinomas (Figure 1A). By feeding these mice with a tamoxifen diet for 2 weeks, we were able to efficiently remove the floxed Pik3ca allele (Figure S1A available online), leaving only one Pik3caWT or Pik3caRBD allele expressed in these mice.
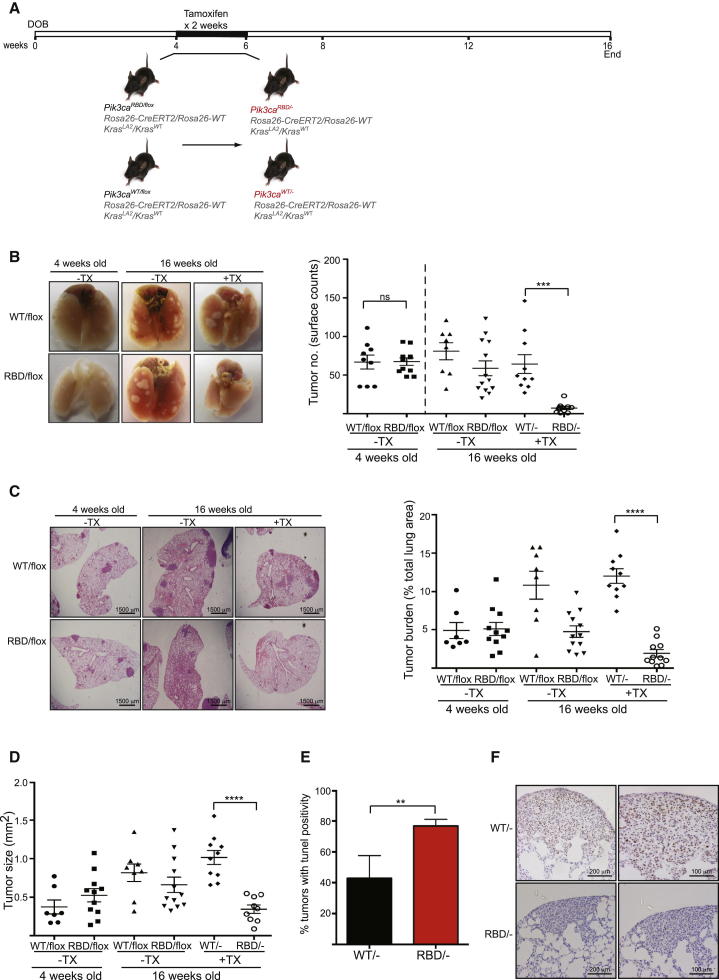
Figure 1.
Expression of p110α-RBD Induces Tumor Regression in Early-Stage Tumors
(A) Schematic representation of experimental conditions.
(B) Representative images of 4-week-old and 16-week-old mouse lungs treated and untreated with tamoxifen (TX). Graph showing tumor number on the pleural surface of lungs. Four-week-old group: Pik3caWT/flox n = 7 mice, Pik3caRBD/flox n = 11; 16-week-old group: Pik3caWT/flox n = 9, Pik3caRBD/flox n = 12, Pik3caWT/- n = 10, Pik3caRBD/- n = 11 mice.
(C) Representative H&E-stained lung sections from 4-week-old and 16-week-old mice. Representation of tumor burden (tumor area as a percentage of total lung area) in 4-week-old mice and in 16-week-old mice 12 weeks after tamoxifen treatment.
(D) Representation of average tumor size from the same group of mice mentioned above.
(E) Quantification of TUNEL positive tumors in 16 weeks old Pik3caRBD/− and Pik3caWT/− mice.
(F) Analysis of the proliferative state of Pik3caWT/− and Pik3caRBD/− tumors using phospho-histone H3 staining.
Error bars indicate mean ± SEM (significance using Student’s t test: ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). See also Figure S1.
Initially, we set out to determine the effect of removing the ability of endogenous p110α to interact with endogenous RAS on RAS-driven tumors at the earliest possible stages, attempting to mimic as closely as possible the constitutive Pik3caRBD mutation knockin reported previously (Gupta et al., 2007). In this we were limited by the fact that tamoxifen treatment of Rosa26-CreERT2 mice in utero or prior to weaning has been associated with Cre recombinase-mediated genotoxicity (Schmidt-Supprian and Rajewsky, 2007). We therefore treated 4-week-old mice with tamoxifen (Figure 1A), an age at which some small tumors had already formed, thus switching the mice to express Pik3caRBD alone (Pik3caRBD/−). We could verify an efficient removal of the floxed allele after the 2-week treatment not only in the tumors, but also in several other tissues (Figure S1A). The number of tumors present on the surface of the lungs of these mice was determined at 16 weeks of age. The number of tumors in Pik3caRBD/− mice was strikingly reduced when compared to their control counterparts, indicating that suppression of RAS interaction with p110α greatly impairs tumor growth and maintenance at this stage (Figure 1B). We also observed a decrease in tumor number in Pik3caRBD/− mice at the end of the experiment when compared to Pik3caRBD/flox mice at 4 weeks of age prior to the beginning of tamoxifen treatment, thus suggesting tumor regression in Pik3caRBD/− mice and not just slower tumor development.
These results were confirmed by histopathological analyses performed on these samples (Figures 1C, 1D, and S1B). Tumor burden, number, and size were greatly reduced in Pik3caRBD/− tumors compared to the Pik3caWT/−, Pik3caRBD/flox, or Pik3caWT/flox tumors. We observed some decrease of the tumor burden in Pik3caRBD/flox mice compared to Pik3caWT/flox mice in the absence of tamoxifen treatment (see below). There was, however, no difference in tumor burden in the heterozygous Pik3caWT/flox mice without (WT/flox) and with (WT/−) tamoxifen treatment, suggesting that Pik3ca gene copy number does not have a major effect on tumor growth at this stage.
We also determined tumor grade according to standard histopathological characteristics (Jackson et al., 2005) and found no differences in the existing tumors between Pik3caRBD/− mice and Pik3caWT/− mice (Figure S1C). Immunohistochemistry staining showed a decrease in phospho-ERK (p-ERK) and p-S6 in Pik3caRBD/− tumors (Figure S1D). Furthermore, the levels of cell proliferation and apoptosis were studied in the tumors of these animals. As shown in Figure 1E, TUNEL staining revealed an increase in the number of tumors displaying apoptosis in Pik3caRBD/− animals when compared to the Pik3caWT/−. The proliferative rates in the tumors in the two backgrounds were also different, as measured by phospho-histone H3 staining for mitotic cells. We could see a higher number of cells stained in the Pik3caWT/− tumors than in the Pik3caRBD/− tumors (Figure 1F), so it is likely that the difference in the sizes of the tumors in the wild-type and the PI3-kinase mutant backgrounds is at least in part due to both elevated rates of cell death and decreased proliferation in the absence of direct RAS binding to p110α.
Previous results in our lab using the constitutive knockin Pik3caRBD mice had shown no differences in tumor formation between wild-type (WT) and heterozygous Pik3caRBD/WT mice, suggesting that the Pik3caRBD allele is functionally recessive to the Pik3caWT allele. However, our results here (Figure 1C) showed some difference in overall tumor burden between Pik3caWT/flox and Pik3caRBD/flox mice. To investigate this further, we studied tumor formation in mice of the same genotype except lacking the Rosa26-CreERT2 allele. In this case, we could not find any significant difference in either tumor number or size, suggesting that the differences found in our Pik3caWT/flox and Pik3caRBD/flox mice could be due to some degree of Cre recombinase activity in Rosa26-CreERT2 mice even in the absence of tamoxifen treatment (Figures S1E and S1F).
Removal of RAS Interaction with PI3-Kinase p110α in Established Tumors Induces Partial Tumor Regression
While the previous experiments demonstrate that the interaction between p110α and RAS plays a critical role in early-stage tumor development and maintenance, the very small size of lung tumors in 4-week-old KrasLA2 mice made it impossible to follow single tumors longitudinally, thus making it hard to distinguish between changes in tumor development (the appearance of new tumors) and changes in tumor maintenance (the persistence of preexisting tumors). We therefore studied the effect of Pik3caRBD expression in more established tumors in adult mice. To address this, we treated 10-week-old mice with tamoxifen and used micro-computed tomography (micro-CT) analysis to monitor the growth of individual tumors over a 6-week period following the start of treatment (Figure 2A). Efficient removal of the floxed Pik3ca allele was observed soon after treatment and at the end of the experiment (Figure S2A). We first monitored tumor growth in the absence of tamoxifen and found that, while individual tumor growth was variable, both Pik3caWT/flox and Pik3caRBD/flox tumors consistently increased in volume over this period (Figure 2B). In contrast, exclusive expression of Pik3caRBD/− following tamoxifen treatment led to partial regression of tumors (Figure 2B). Furthermore, to investigate whether activation of Cre recombinase may by itself have an impact on tumor growth, mice that had Rosa26-CreERT2 but no loxP sites flanking Pik3ca were also treated with tamoxifen. We found that these tumors continued to grow following treatment, and thus the effects observed above are directly dependent on p110α and are not due to Cre recombinase-associated toxicity (Figures 2B, green lines, and S2B).
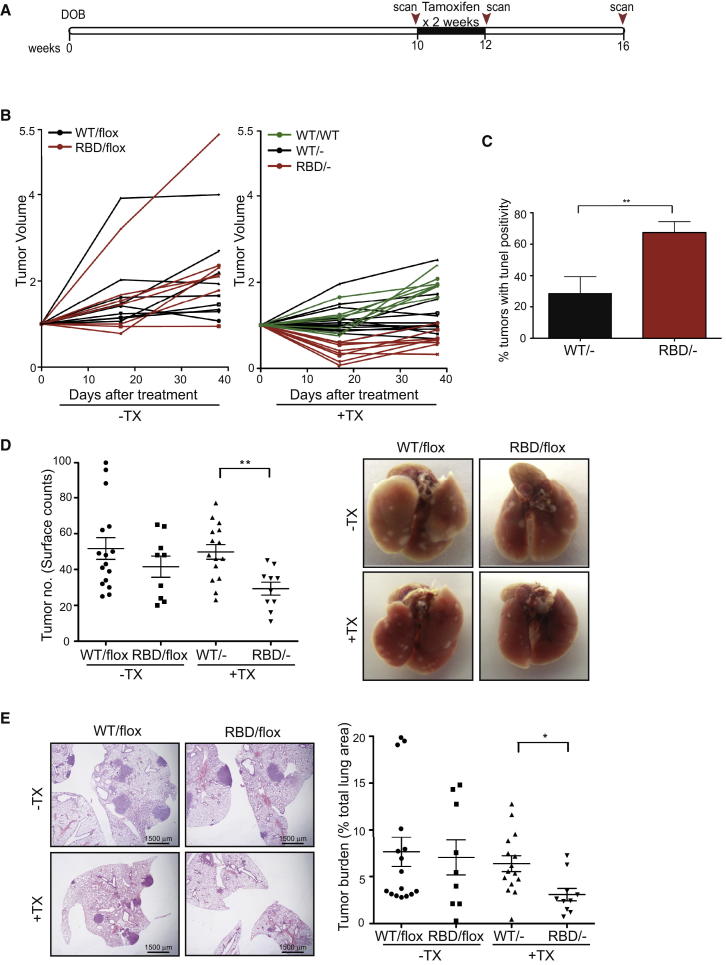
Figure 2.
Expression of p110α-RBD in Established Tumors Reduces Tumor Burden
(A) Schematic representation of mouse treatment and micro-CT scanning intervals.
(B) Ten-week-old mice were left untreated or were treated with tamoxifen for 2 weeks and the effect on tumor volume was assessed by micro-CT analysis over a 6-week period. Tumor volumes are plotted relative to the initial volume at the start of treatment. Black lines represent tumors from Pik3caWT/− mice and red lines represent tumors from Pik3caRBD/− mice. Green lines represent tumors from tamoxifen-treated Pik3caWT/WT mice that express Cre recombinase following treatment but do not have loxP sites flanking the Pik3ca gene. Pik3caWT/flox n = 4 mice, Pik3caRBD/flox n = 4, Pik3caWT/- n = 6, Pik3caRBD/- n = 4, Pik3caWT/WT n = 3 mice.
(C) Quantification of TUNEL-positive tumors following tamoxifen treatment for 11 days. Pik3caWT/− n = 5 mice, Pik3caRBD/− n = 7 mice
(D) Quantification of tumor multiplicity on the pleural surface of lungs. Representative images of lungs from Pik3caWT/flox, Pik3caRBD/flox, Pik3caWT/−, and Pik3caRBD/− mice at 16 weeks of age are shown.
(E) Quantification of tumor burden (tumor area as a percentage of total lung area) in mice 6 weeks after commencement of tamoxifen treatment. Representative images of histological sections stained with H&E in Pik3caWT/flox, Pik3caRBD/flox, Pik3caWT/−, and Pik3caRBD/− mice at 16 weeks of age are shown.
Error bars indicate mean ± SEM (significance using Student’s t test: ∗p < 0.05 ∗∗p < 0.01). See also Figure S2.
As initial tumor regression was evident following tamoxifen treatment in Pik3caRBD/− mice, we carried out TUNEL staining on samples generated 10 days after the commencement of tamoxifen treatment. This revealed that cell death occurred in many of the mutant tumors following tamoxifen treatment, although widespread involution of tumors was not evident (Figure 2C). The effect of mutant expression on tumors was also evident using histological analysis of lungs (Figures 2D, 2E, and S2C). The lungs in Pik3caRBD/− mice had fewer tumors present and the overall tumor burden in these animals was reduced. In addition, examination of downstream signaling revealed that while the level of AKT phosphorylation varied in individual tumors, mutant tumors displayed lower activation of AKT than WT tumors (Figure S2D).
Removal of RAS Interaction with PI3-Kinase p110α Inhibits Tumor Progression
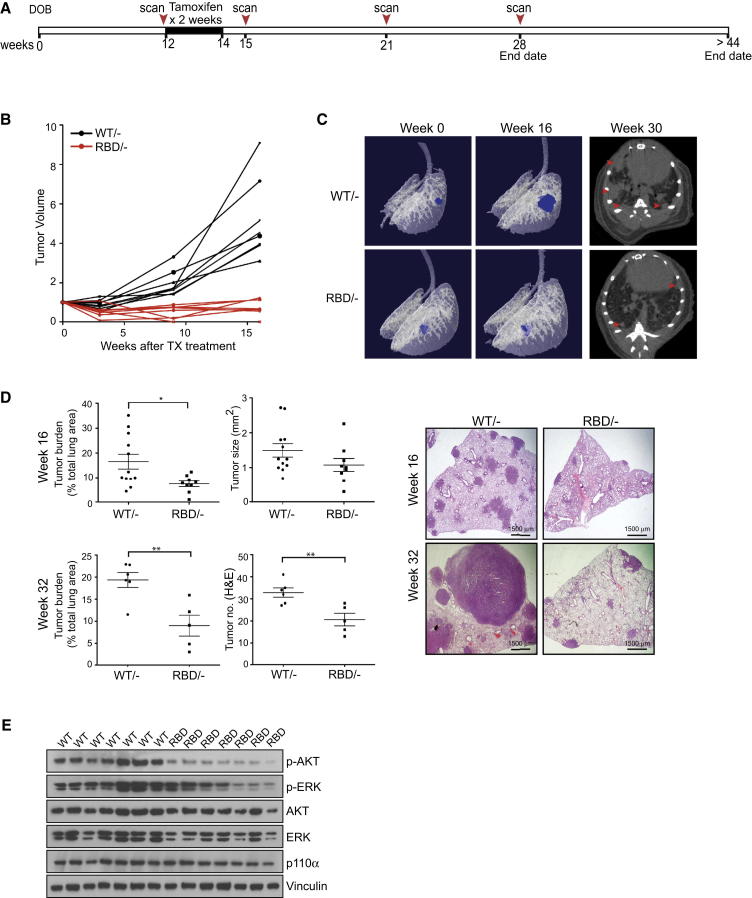
In order to determine the more long-term consequences of Pik3caRBD expression on tumor development, we followed tumor growth for 16–32 weeks after tamoxifen treatment (Figure 3A). Examination of individual tumor growth revealed that while Pik3caWT/− tumors recovered after the initial tamoxifen treatment and showed robust tumor expansion over a 4-month period, the Pik3caRBD/- tumors remained static and did not progress further (Figures 3B and S3A). This was particularly clear in the 3D tomographs of lungs from treated mice (Figure 3C). Over time, Pik3caWT/− tumors accumulated in the lungs, with some tumors merging; however, tumor multiplicity remained low in the Pik3caRBD/− mice (Figure 3C, right). Interestingly, the tumor burdens in both Pik3caWT/− and Pik3caRBD/− mice were lower than in Pik3caWT/flox and Pik3caRBD/flox mice, indicating that some gene dosage effects may be occurring (Figure S3B). Examination of p110α levels by western blot revealed that a decrease in p110α expression occurs during tamoxifen treatment, which may transiently affect growth in Pik3caWT/− tumors (Figure S3C). Our results demonstrate that in established KrasLA2-driven tumors, the PI3-kinase pathway continues to be an important mediator of tumor progression. Indeed, at these later time points, tumor burden was significantly decreased in Pik3caRBD/− mice versus Pik3caWT/− mice and the difference in tumor size became more evident as the Pik3caWT/− tumors continued to grow (Figures 3D and S3D). Furthermore, activation of the AKT pathway downstream of PI3-kinase was impaired in Pik3caRBD/− mice, indicating that AKT signaling downstream of RAS continues to be disrupted (Figure 3E). We also utilized standard histological grading to look at tumor progression (Figure S3E). While overall tumor number was lower in Pik3caRBD/− mice, the incidence of higher-grade tumors was also reduced in these mice, with few grade 3 and no grade 4 adenocarcinomas detected in mutant mice 16 weeks after treatment (Figure S3F). However, at later stages of the experiment, some regrowth of mutant tumors was observed (Figure S3G). Examination of these tumors revealed the presence of unrecombined Pik3ca in mutant tumors. This indicates that over extended periods of time, a minority of cells that escaped Cre-mediated recombination during tamoxifen treatment may proliferate and restore tumor growth in Pik3caRBD/− mice (Figure S3G).
Figure 3.
p110α-RBD Mutation Inhibits Tumor Progression
(A) Schematic representation of mouse treatment and micro-CT scanning intervals.
(B) The long-term effect of Pik3caRBD expression on tumor volume was assessed using micro-CT analysis. Lungs were scanned before tamoxifen treatment and at 3, 9, and 16 weeks after commencement of treatment. Tumor volumes are plotted relative to the initial volume at the start of treatment. Black lines represent tumors from Pik3caWT/− mice and red lines represent tumors from Pik3caRBD/− mice. Pik3caWT/− n = 3 mice, Pik3caRBD/− n = 3 mice.
(C) Representative tomographs from micro-CT analysis of Pik3caWT/− and Pik3caRBD/− mice before tamoxifen treatment and 16 weeks later. Representative micro-CT images from each genotype 30 weeks after treatment are shown on the right.
(D) Histological quantification of tumor burden, tumor size, and tumor number either 16 weeks or >32 weeks after the commencement of tamoxifen treatment. Representative histological images of lungs from Pik3caWT/− and Pik3caRBD/− mice are shown.
(E) Western blot analysis of AKT phosphorylation in tumors extracted from the lungs of Pik3caWT/− and Pik3caRBD/− mice 15 weeks after commencement of tamoxifen treatment.
Error bars indicate mean ± SEM (significance using Student’s t test: ∗p < 0.05 ∗∗p < 0.01). See also Figure S3.
We were also interested in the impact of Pik3caRBD expression on immune cell infiltration into the lungs. To look at this, we stained lung samples for markers of different immune cell subtypes. While histological staining showed no dramatic effect of Pik3caRBD on neutrophils, B cells, or T cells 16 weeks after treatment, we did see a marked decrease in the number of F4/80+ macrophages in the lungs (data not shown; Figure S3H). Tumor-associated macrophages have been observed previously in mutant Kras-induced lung tumors in mice and have broadly been associated with tumor progression (De Palma and Lewis, 2013; Ji et al., 2006; Pollard, 2004). This suggests that in addition to direct inhibition of tumor growth, disruption of RAS signaling to p110α may also affect crosstalk between the tumor and nontumor host cells.
Tumor Cell-Autonomous Regression on Removal of RAS Interaction with PI3-Kinase p110α
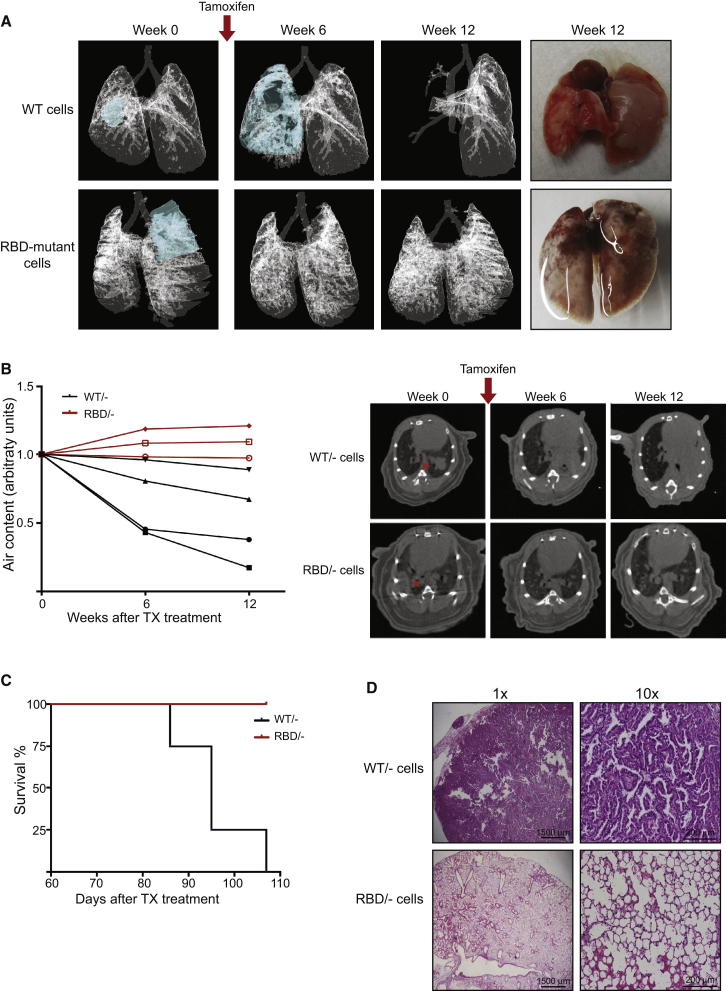
While the results above indicate a defect in tumor growth and maintenance in Pik3caRBD/− mice, it is possible that this defect is not entirely tumor cell autonomous. The Pik3caRBD protein is present not only in the tumor cells, but also surrounding host cells, including stromal, endothelial, and immune cells, any of which could contribute to the observed tumor regression or stasis. In order to investigate this, we derived lung tumor epithelial cells from 7 to 9 months old KrasLA2Rosa26-CreERT2 mice that were either Pik3caRBD/flox or Pik3caWT/flox. These animals had not been exposed to tamoxifen, so the floxed Pik3ca allele in these cells remained unrecombined. Tumor cells were sorted to remove immune and endothelial cells and were delivered intratracheally into the lungs of nude (NuNu) mice. Between 10 and 16 weeks after transplantation, tumors began to be visible by micro-CT in the lungs of the nude mice, with in all cases just one tumor detectable per lung. When tumors were visible by micro-CT, mice were treated with tamoxifen to delete the floxed Pik3ca allele in the tumor cells and the tumor growth was followed over a period of 12 weeks. In mice transplanted with Pik3caWT/flox tumor cells, tumors continued to grow rapidly after tamoxifen treatment, with a marked decrease in air content available in the lung (Figures 4A and 4B). However, in mice bearing Pik3caRBD/flox tumor cells, tumors completely disappeared after tamoxifen treatment and had not regrown by the end of the experiment, with lung air content increasing somewhat over time (Figures 4A and 4B). Furthermore, these effects led to enhanced survival of mice inoculated with Pik3caRBD/flox tumor cells versus Pik3caWT/flox tumor cells (Figure 4C).
Figure 4.
Deficient Tumor Growth in p110α-RBD Mutant Mice Is a Cell-Autonomous Process
(A) 3D models showing the growth of transplanted Pik3caWT/− and Pik3caRBD/− tumor cells in the lung based on micro-CT images. Due to the tumor occupying the complete lobe at 12 weeks after treatment in Pik3caWT/− inoculated lungs, modeling was not possible.
Representative images of Pik3caWT/− and Pik3caRBD/− tumor cell inoculated lungs 14 weeks after tamoxifen treatment are shown in the most right panels.
(B) Mice were scanned 6 and 12 weeks after tamoxifen treatment. The graph represents the normalized air volume in the lungs at the moment of the scanning. Black lines represent air content in NuNu mice bearing Pik3caWT/−-derived tumors and red lines represent air content in mice bearing Pik3caRBD/−-derived tumors.
Representative tomographs from micro-CT analysis of Pik3caWT/−-derived and Pik3caRBD/−-derived tumors are shown at the right.
(C) Representation of lifespan of NuNu mice bearing either Pik3caWT/−-derived or Pik3caRBD/−-derived tumors. NuNu mice containing Pik3caRBD/−-derived tumors were sacrificed when the last mouse from the control group had to be culled because of breathing problems.
(D) Representative images of histological sections stained with H&E in mice inoculated with Pik3caWT/−-derived and Pik3caRBD/−-derived tumor cells 14 weeks after tamoxifen treatment.
Histopathology analysis of the lungs of these mice 12 weeks after tamoxifen treatment showed similar results. NuNu mice containing Pik3caWT/flox tumor cells had large tumors, generally just one occupying most of the lobe, although in several of them there were also small tumor foci in the vicinity of the main one. All the tumors presented a dominant papillary architecture and were usually classified as grade 3. Lungs in those mice explanted with Pik3caRBD/flox cells presented no tumors and had a normal tissue structure (Figure 4D). These results indicate a dependence on tumor cell-autonomous RAS interaction with p110α for tumor maintenance and growth. However, they do not rule out the possibility that the loss of interaction of RAS with p110α in host cells might also contribute to the regression of tumors seen in the autochthonous setting.
Complete Removal of p110α in Established Tumors Similarly Reduces Tumor Burden
Our experiments thus far have revealed the importance of PI3-kinase activation by oncogenic Kras for the progression and continued development of KrasLA2-dependent lung tumors. While the crucial role of RAS-mediated activation of the PI3-kinase pathway during initial tumor development has been clearly demonstrated (Gupta et al., 2007), established tumors may utilize both internal oncogenic RAS and exogenous growth factors to activate this important pathway for continued tumor progression. We were thus interested to investigate whether complete removal of p110α would provide an additional effect on tumor growth in comparison to p110α-RBD expression. To explore this, we treated Pik3caflox/flox mice at 10 weeks of age with tamoxifen and followed individual tumor growth (Figure 5A). Tamoxifen treatment resulted in efficient removal of p110α expression in tumors at the protein level (Figure 5B). In tamoxifen-treated Pik3caflox/flox mice, we observed a partial regression of tumors in a manner similar to Pik3caRBD/− expression (Figure 5C). While the effect of complete removal of p110α expression was slightly more robust, the change in tumor volume at the end of the 6-week period was comparable to that seen with Pik3caRBD/− expression (Figure 5D). Thus, in these tumors p110α activation appears to be largely dependent upon interaction with RAS.
Figure 5.
Complete Removal of p110α in Established Tumors Similarly Reduces Tumor Burden
(A) Schematic representation of mouse treatment and micro-CT scanning intervals.
(B) Western blot analysis of p110α expression in tumors extracted from Pik3caflox/flox and Pik3ca−/− lungs 3 weeks after commencement of tamoxifen treatment.
(C) Ten-week-old mice were left untreated or were treated with tamoxifen for 2 weeks and the effect on tumor volume was assessed by micro-CT analysis over a 6-week period. Black lines represent tumors from Pik3caflox/flox mice and blue lines represent tumors from Pik3ca−/− mice. Pik3caflox/flox n = 3 animals, Pik3ca−/− n = 4 animals.
(D) Change in tumor volume in Pik3caWT/WT, Pik3caWT/RBD, Pik3caWT/−, Pik3caRBD/-, and Pik3ca−/− mice. Ten-week-old mice were left untreated or were treated with tamoxifen for 2 weeks and the effect on tumor volume was assessed by micro-CT analysis over a 6-week period. Tumor volumes are plotted relative to the initial volume at the start of treatment.
(E) Histological quantification of tumor number and tumor burden in mice 6 weeks after commencement of tamoxifen treatment. Representative images of lungs from Pik3caflox/flox and Pik3ca−/− at 16 weeks of age are also shown.
(F) The long-term effect of Pik3ca removal on tumor volume was assessed using micro-CT analysis. Lungs were scanned before tamoxifen treatment and at various intervals up to 12 weeks after the commencement of treatment. Black lines represent tumors from Pik3caflox/flox mice and blue lines represent tumors from Pik3ca−/− mice. Pik3caflox/flox n = 3 animals, Pik3ca−/− n = 3 animals.
(G) Glucose tolerance test of Pik3caflox/flox, Pik3caWT/−, Pik3caRBD/−, and Pik3ca−/− mice 3 weeks after the end of tamoxifen treatment. Mice were injected with glucose (1 g/kg) and blood glucose levels were measured at the indicated time points. Glucose levels are plotted relative to the initial value before injection. Pik3caflox/flox n = 8 animals, Pik3ca−/− n = 11 animals, Pik3caWT/− n = 6 animals, Pik3caRBD/− n = 8 animals.
Error bars indicate mean ± SEM (significance using Student’s t test: ∗p < 0.05). See also Figure S4.
Histological analysis of Pik3ca−/− mice also showed a clear decrease in the number of tumors present and the overall tumor burden, while tumor size was not significantly changed (Figures 5E and S4A). Furthermore, ablation of p110α expression in LKR13 cells established from lung tumors in the KrasLA2 mouse model resulted in decreased proliferation of these cells (Figure S4B). This was accompanied by a reduction in downstream AKT activation. In contrast, ablation of p110β had little effect on proliferation or AKT signaling, indicating that these cells are primarily dependent on the p110α isoform of PI3-kinase. Analysis of the long-term effects on tumor growth after tamoxifen treatment in Pik3caflox/flox mice also demonstrated that tumor progression was inhibited (Figure 5F). However, regrowth of tumors occurred more rapidly in these mice than in Pik3caRBD/− mice. Some residual expression of p110α was found in remaining tumors, indicating that incomplete recombination in some cells may allow re-establishment of PI3-kinase signaling and proliferation in tumors (Figure S4C).
While both p110α-RBD expression and p110α depletion have a clear effect on oncogenic Kras-driven tumor advancement, these two genetic alterations may have different effects on PI3-kinase signaling in nontumorigenic cells. This will be dictated by the extent of RAS involvement during growth factor-mediated activation of PI3-kinase signaling. Previous studies exploring the effect of p110α deletion/blockade on insulin signaling revealed that this isoform of PI3-kinase is a key mediator of metabolic responses to insulin (Knight et al., 2006; Sopasakis et al., 2010). We performed a glucose tolerance test on untreated mice and tamoxifen-treated Pik3caWT/flox, Pik3caRBD/flox, and Pik3caflox/flox mice to investigate whether insulin-mediated removal of glucose from the blood was affected by genetic alteration of Pik3ca. We found that deletion of Pik3ca led to a greater impairment in glucose removal from the blood, indicating that nontumorigenic functions of p110α may be affected more by removal of p110α compared to expression of p110α-RBD (Figure 5G). Furthermore, the health of Pik3ca−/− mice was more adversely affected than Pik3caRBD/− mice, with significantly more mortality occurring within 4 weeks of the start of treatment (Figure S4D). These results suggest that disruption of RAS and p110α interaction may target tumor cells more specifically and may be accompanied by fewer side effects associated with global inhibition of p110α PI3-kinase function.
Combined PI3-Kinase Pathway Inhibition with MEK Inhibition Causes Enhanced Tumor Regression
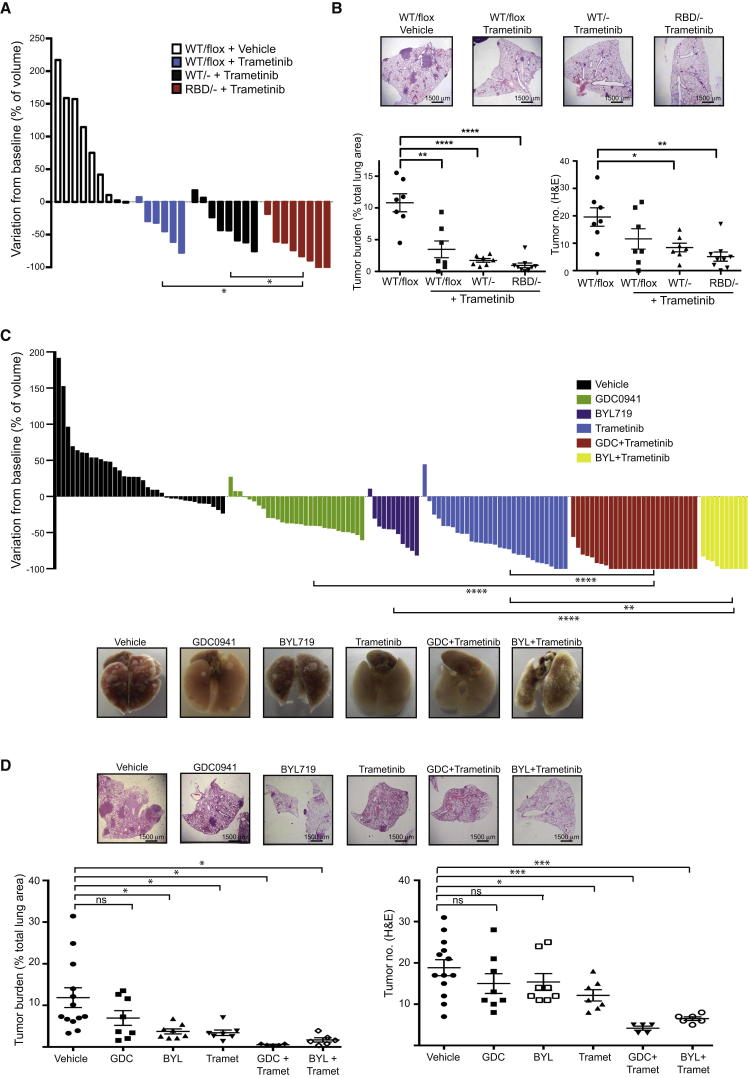
We have found that expression of p110α-RBD caused partial regression in established tumors, with long-term tumor stasis. However, with this mutant, we have targeted only one of the major pathways downstream of RAS, and continued activation of other pathways may provide sufficient survival signals to maintain tumor integrity, at least in part. To investigate whether the ERK/mitogen-activated protein kinase (MAPK) pathway was facilitating the survival of Pik3caRBD;KrasLA2 tumors, we decided to target this signaling pathway in conjunction with disruption of PI3-kinase signaling. We treated adult KrasLA2;Rosa26-CreERT2; Pik3caRBD/flox or KrasLA2;Rosa26-CreERT2; Pik3caWT/flox mice with tamoxifen for 2 weeks and then treated with the MEK inhibitor trametinib for a further 4 weeks, using micro-CT imaging to monitor the effect on tumors. Treatment with trametinib promoted partial tumor regression in Pik3caWT/− mice, but the effects were significantly stronger in Pik3caRBD/− mice, with complete regression of some tumors (Figures 6A, S5A, and S5B). Similar results were found when mice were treated with the lung cancer chemotherapy combination carboplatin plus paclitaxel, with Pik3caRBD/− mice showing a stronger response than Pik3caWT/− animals (Figure S5A). We confirmed that trametinib efficiently inhibited MEK by assessing the levels of p-ERK in the lungs (Figure S5C). Histological analysis also revealed a dramatic reduction of tumor burden in these mice (Figures 6B and S5D). There was a significant decrease in tumor number as many of the lesions regressed and tumors that remained were small and dispersed unlike their larger more compact counterparts in untreated mice (Figure S5E). Indeed, untreated tumors were consistently higher grade than those in the combination treatment (Figure S5F). Tumor burden was also reduced with combined tamoxifen and trametinib treatment of Pik3caWT/flox mice, likely due in some part to the initial impairment of tumor growth that was observed following removal of a single allele of Pik3caWT (Figures 2B and S5A).
Figure 6.
Combined PI3-Kinase Inhibition with MEK Inhibition Induces Tumor Regression
(A) Waterfall plot showing tumor response in 10-week-old mice that were treated with tamoxifen for 2 weeks, left untreated for 5 days, and then treated with trametinib for 4 weeks. Each bar represents one tumor and response is shown relative to initial tumor volume before tamoxifen treatment.
(B) Representative histological images of lungs from mice, histological quantifications of tumor burden, and tumor number in mice treated as described in (A) are shown.
(C) KrasLA2 mice were treated with GDC0941 (100 mg/kg/day; green bars), BYL719 (50 mg/kg/day; purple bars), trametinib (3 mg/kg/day; blue bars), the combination of GDC0941 or BYL719 with trametinib (full dose for each drug; red and yellow bars, respectively), or vehicle alone (black bars). The waterfall plot represents response on tumor growth after 4 weeks treatment. Each bar represents one tumor and response is shown relative to initial tumor volume before tamoxifen treatment. Representative images of lungs from mice on each treatment are also shown.
(D) H&E staining, tumor burden representation, and tumor number assessment from lungs treated with the drugs described in (C).
Error bars indicate mean ± SEM (significance using Student’s t test: ∗p < 0.05 ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). See also Figure S5.
Next, we aimed to compare these results with pharmacological inhibition of PI3-kinase in a similar Kras-driven tumorigenesis background. To do so, we treated KrasLA2 mice with GDC0941, an orally bioavailable inhibitor of all class I PI3-kinases, BYL719, a p110α isoform specific inhibitor, and the combination of GDC0941 or BYL719 and trametinib. We observed a modest decrease in tumor growth in mice treated with GDC0941 or BYL719 alone (Figure 6C). Interestingly, treatment of these mice with GDC0941 or BYL719 alone gave similar tumor responses, suggesting that the decrease in tumor growth is mainly achieved by inhibition of p110α isoform. When KrasLA2 mice were treated with trametinib together with GDC0941, or with BYL719, a strong tumor regression response was obtained, with an almost total disappearance of the tumors as evaluated by micro-CT scanning (Figure 6C). However, the toxicity was higher in the combined treatments, and treatment had to be stopped in the final week of the 5-week course in about half of the mice in these cohorts. The GDC0941 and trametinib combination tumor regression response was significantly stronger than that obtained with carboplatin plus paclitaxel or GDC0941 combined with carboplatin plus paclitaxel (Figure S5G). These results were confirmed in hematoxylin and eosin (H&E) staining where we could see an almost complete disappearance of the tumors in lungs from mice treated with the combined trametinib with GDC0941 or with BYL719 therapy (Figures 6D and S5H–S5J). Finally, we checked the efficiency of the GDC0941 and trametinib treatments in hitting their targets by assessing p-ERK and p-AKT levels in tumors of mice treated daily by these drugs. Both AKT and ERK phosphorylation were inhibited in tumors treated with their respective pathway inhibitors, either alone or in combination (Figure S5K).
Discussion
In these studies, we have attempted to assess the ongoing requirements of preexisting mutant Kras-induced lung tumors for direct interaction of RAS with p110α and to determine whether they differ from the previously established almost absolute requirement for this interaction in the de novo development of Kras-driven lung tumors (Gupta et al., 2007). We show here that genetically blocking the interaction of RAS with p110α causes partial regression of tumors, followed by long-term tumor stasis, although by itself it is not sufficient to cause complete regression of established tumors in the autochthonous setting. This suggests that eradication of existing tumors is harder to achieve than prevention of the formation of new tumors, with the requirements for tumor maintenance being less stringent than for tumor development. However, combination targeting of RAS’s interaction with p110α with inhibition of the RAF/ERK pathway at MEK causes much improved tumor regressions. While RAS-induced activation of PI3-kinase is clearly important in the continued growth of Ras mutant lung cancer in this system, we did not see a clear-cut pathway dependency for tumor maintenance as has been reported by others (Lim and Counter, 2005), but rather contributions of both PI3-kinase and RAF/MEK/ERK pathways. Although the oncogenic role of RAS in the promotion of lung cancer has been known for many years, treatment strategies that effectively target RAS mutant tumors have yet to be established. In contrast to the progress observed with epidermal growth factor receptor (EGFR) or activin receptor-like kinase (ALK) inhibitors in lung cancers with alterations in those driver oncogenes, specific inhibition of RAS mutant cancers has been difficult to achieve (Maemondo et al., 2010; Shaw et al., 2013). Our results demonstrate that RAS-induced PI3-kinase activation is clearly important in these tumors in a mouse model of Kras-driven lung adenocarcinoma. The PI3-kinase pathway thus provides an attractive therapeutic target, with combination of PI3-kinase inhibition with parallel pathway inhibitors or chemotherapeutic drugs likely to provide the greatest benefit.
The value of testing the therapeutic response of Kras mutant lung cancers in mice to the treatment of human disease is clearly highly dependent on how accurately the genetically engineered mouse model used reflects human KRAS mutant lung cancer in the clinic. Several mouse models of Kras-driven lung cancer have been developed over the past two decades (Kim et al., 2005). The model that we use in this study, KrasLA2 (Johnson et al., 2001), is designed to have sporadic expression in single cells of G12D mutant Kras off its own promoter triggered by spontaneous intrachromosomal recombination. While the mutant Kras protein is expressed in a few cells in all tissues, tumor development is seen only in the lung at high penetrance, with occasional thymic lymphomas also occurring. In another heavily used model, KrasLSL-G12D (Jackson et al., 2001), expression of mutant Kras is induced by local introduction of Cre recombinase activity in the lung using an adenoviral vector, with the number of cells expressing mutant Kras being dependent on the viral titer applied. The KrasLA2 model recapitulates many of the features of human Kras mutant lung cancer, and this model has played an important role in extending our knowledge of this disease (Hatley et al., 2010; Hollander et al., 2011; Takahashi et al., 2010). Pathological progression of tumors in these mice from hyperplasia to adenoma and then adenocarcinoma over time phenocopies the progression of human lung tumors and permits establishment of a tumor microenvironment in the lungs. Furthermore, the partial response seen here of lung tumors in KrasLA2 mice to the current standard of care chemotherapies utilized against human lung cancer suggests that this model does to some degree reflect the therapeutic response of human Kras mutant lung cancers. Extending the observations described in our study to more complex Kras/p53 or Kras/Lbk1 mouse models at a later stage may also provide additional insights into human KRAS mutant lung cancer biology and treatment.
The earliest point at which Pik3ca gene recombination could be induced without nonspecific Cre recombinase toxicity was postweaning, at 4 weeks of age, at which time small lung tumors are readily detectable in KrasLA2 mice by histology or visual examination of the lung surface, although not by micro-CT scanning. Although these tumors cannot be followed longitudinally, it appears that they undergo more profound regressions than larger tumors treated at 10 weeks. It is possible that larger tumors acquire additional mechanisms in conjunction with direct input from RAS for maintaining PI3-kinase pathway activity. There thus appears to be some evidence for a progressive decrease in the degree of dependency on direct interaction of RAS with p110α as tumors become more established.
One concern with the approach used here to study the role of the RAS binding domain of p110α in vivo is the effect of changing gene dosage of Pik3ca following Cre recombinase-mediated deletion of a heterozygous floxed allele. For example, Pik3caRBD expression and reduction of the WT level may both contribute to the tumor regression observed in Pik3caRBD/− mice following tamoxifen treatment. However, this has been controlled for in all experiments with comparisons made between a single RBD mutant and a single WT allele of Pik3ca, and we consistently observed enhanced antitumor effects with Pik3caRBD expression. When comparison was made between mice in which WT Pik3ca gene dosage was changed from two to one, some decrease in tumor growth rate, but never actual tumor regression, was seen.
In order to distinguish tumor cell-intrinsic requirements for RAS’s interaction with p110α from effects involving nontumor cells of the host, such as stroma, endothelial, and immune cells, we carried out orthotopic transplantation experiments using tumor cells from engineered mice introduced into the lungs of animals with normal PI3-kinase signaling. These showed that a major component of the inhibitory effect of removing the RAS and p110α interaction on tumor fate could be recapitulated when the interaction is prevented in the tumor cells alone. We cannot rule out the possibility that changes in host cell signaling also contribute, but there is clearly a major tumor cell-autonomous effect. The fact that tumor regression occurs in nude mice indicates that this can occur without the adaptive immune system, although again it is not possible to exclude a contributing role. It is also apparent that transplanted tumors actually show much more profound regressions than autochthonous tumors when the RAS and p110α interaction is blocked. One might speculate that the tumor niche established on transplantation provides a less robustly trophic environment for the tumor cells than the niche that evolves during the extended evolution of an autochthonous tumor.
Comparison of the effect on Kras-driven autochthonous tumors of total deletion of p110α with the removal of the ability of p110α to interact with RAS reveals similar levels of tumor regression and stasis. This indicates that in this model of lung tumors driven by activated Kras, the direct binding of RAS to p110α is a critical component of the regulation of this PI3-kinase isoform. However, it is clear that ablating only the RAS interaction with p110α has less systemic toxicity for the mice than total p110α loss, as exemplified by less disruption of glucose homeostasis and also better general health status. It is noticeable, however, that ubiquitous loss of p110α in adult mice does not lead to very major effects on mouse viability when compared with p110α loss or inactivation in developing embryos (Bi et al., 1999; Foukas et al., 2006), possibly reflecting some redundancy between PI3-kinase isoforms, as well as inevitably less than 100% deletion of Pik3ca on Cre activation. It is possible that a pharmacological strategy that targeted RAS interaction with p110α may have fewer adverse effects than targeting p110α lipid kinase activity, but it may still be effective in opposing tumor progression. Recently, some progress has been made in identifying small molecules that bind directly to RAS (Maurer et al., 2012; Shima et al., 2013; Sun et al., 2012), and these might eventually lead to drugs that would be able to block the interaction of RAS with downstream effector enzymes. At present, however, the relatively low potency of the compounds identified, which is low μM at best, has precluded assessment of their efficacy in the Kras lung cancer mouse model.
Another likely advantage of direct RAS effector interaction site targeted drugs would be that they would also be expected to inhibit other RAS effector pathways such as RAF/MEK/ERK, which are activated from the same region of RAS. It has long been clear that inhibition of MEK and PI3-kinase together promotes death of RAS mutant cancer cells (Engelman et al., 2008), although this approach has significant toxicities and may not show enhanced selectivity for the RAS mutant phenotype relative to cells with wild-type RAS (Molina-Arcas et al., 2013). Combining pharmacological inhibition of MEK together with genetic blockade of RAS interaction with p110α also considerably improved tumor clearance in the model system used here, as did drug-mediated inhibition of both MEK and type I PI3-kinases. Use of a p110α isoform-specific PI3-kinase inhibitor had similar effects to a pan type I PI3-kinase inhibitor, reinforcing the view that p110α is the critical PI3-kinase isoform in this setting. We now have evidence that RAS does not control p110β through direct interaction (Fritsch et al., 2013), so it is unlikely that inhibition of this ubiquitous isoform will be helpful in RAS-driven cancer. The other type I PI3-kinase isoforms, p110γ and p110δ, are unlikely to be expressed at major levels in the epithelial tumor cells studied here, although their inhibition could play an important role in the interaction of the tumor with host cells, especially those of the innate and adaptive immune systems.
Achieving effective targeting of the multiple signaling pathways driven by RAS through direct blockade of the effector-binding region of RAS remains a tantalizing, if distant, goal in cancer drug discovery. The possibility that drugs might be made that are specific for mutant, rather than wild-type, RAS proteins could also add greatly to their therapeutic potential. The results reported here suggest that such an approach could show great promise in the treatment of RAS mutant cancers if the challenge of finding effective RAS-effector protein-protein interaction inhibitors could be met.
Experimental Procedures
Mouse Model Generation
The final mouse model was generated by breeding of four different mouse strains: Pik3caRBD (Gupta et al., 2007), KrasLA2 (Johnson et al., 2001), C57BL/6-Gt(Rosa)26Sortm9(cre/ESR1)Arte (Taconic Artemis Gmbh; referred to here as Rosa26-CreERT2), and Pik3caflox/flox mice (Zhao et al., 2006). All animal experiments were approved by the CRUK London Research Institute Animal Ethics Committee and conformed with UK Home Office regulations under The Animals (Scientific Procedures) Act 1986 including Amendment Regulations 2012.
Antibodies
Total ERK, p-ERK (Thr202/Tyr204 -E10) monoclonal, total AKT, pAKT (Ser473), p-S6, p110α, and p110β antibodies were all purchased from Cell Signaling Technology. Vinculin antibody was purchased from Sigma. p-PRAS40 (Thr246) was purchased from Invitrogen. For immunohistochemistry, F4/80 antibody was purchased from eBioscience.
Reagents
Tamoxifen diet (400 mg/kg) was obtained from Harlan (Teklad Lab animal diets). D(+) glucose was obtained from Sigma. Small interfering RNA pools were obtained from Dharmacon. Trametinib (GSK1120212) and BYL719 were obtained from Active Biochem. GDC0941 and paclitaxel were purchased from LC Laboratories and Carboplatin from Selleck Chemicals.
In Vivo Tamoxifen and Drug Treatments
Mice were treated with tamoxifen food pellets for 2 weeks and returned to normal diet for the duration of the experiment. To confirm that Cre-mediated excision of the floxed allele occurred, DNA was extracted from tumors, lung, and other tissue samples using Extract-N-Amp Tissue PCR kit (Sigma). Genotyping PCRs for the Pik3ca and Pik3caflox alleles was then carried out using the following primers: forward 5′-CTGTGTAGCCTAGTTTAGAGCAACCATCTA-3′, reverse 5′-CCTCTCTGAACAGTTCATGTTTGATGGTGA-3′.
For therapeutic drug treatments, drugs were given by oral gavage daily with vehicle, trametinib (3 mg/kg/day), GDC0941 (100 mg/kg/day), or BYL719 (50 mg/kg/day) for 5 weeks. Trametinib and GDC0941 were dissolved as described previously (Gilmartin et al., 2011). Paclitaxel (10 mg/kg/day) and carboplatin (50 mg/kg/day) were administered by intraperitoneal injections twice a week, and both drugs were dissolved in PBS.
Histology and Analysis of Tumor Burden
Mice were sacrificed and the lung tissue was immediately removed and fixed overnight in 10% neutral buffered formalin. The lungs were then transferred to 70% ethanol and processed for paraffin embedding. Tissue sections were cut at 4 μm and were stained with H&E or were immunostained with various antibodies. Lung and tumor area quantifications were carried out on H&E-stained slides. Pictures of each lung lobe were taken on a Nikon Eclipse 90i microscope with the 1× objective. Lung and tumor area were measured using NIS Elements 3.0 SP7 software (Nikon Instruments B.V. Europe). Tumor grading was carried out by a pathologist and tumors were classified according to standard histopathological grading (Jackson et al., 2005).
Statistical Analysis
Data are presented as mean ± SEM. Significance was determined with GraphPad Prism 5 software using the Student’s t test unless stated otherwise.
Acknowledgments
We thank Jean Zhao for providing Pik3caflox/flox mice and Miriam Molina-Arcas, Ralph Fritsch, and David Hancock for help and advice. We thank LRI Core Technology Facilities, including Biological Resources. C.S. was supported by an EMBO long-term fellowship. This work was funded by Cancer Research UK, the European Union Framework Programme 7 grant “LUNGTARGET,” the European Research Commission Advanced grant “RASTARGET,” the NC3Rs grant NC.K000500.1, and the Association for International Cancer Research grant 13-0142.
Published: November 11, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.ccr.2013.09.012.
Supplemental Information
References
- Bi L., Okabe I., Bernard D.J., Wynshaw-Boris A., Nussbaum R.L. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J. Biol. Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- Cully M., You H., Levine A.J., Mak T.W. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- De Palma M., Lewis C.E. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23:277–286. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Engelman J.A., Chen L., Tan X., Crosby K., Guimaraes A.R., Upadhyay R., Maira M., McNamara K., Perera S.A., Song Y. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foukas L.C., Claret M., Pearce W., Okkenhaug K., Meek S., Peskett E., Sancho S., Smith A.J., Withers D.J., Vanhaesebroeck B. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- Fritsch R., de Krijger I., Fritsch K., George R., Reason B., Kumar M.S., Diefenbacher M., Stamp G., Downward J. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153:1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin A.G., Bleam M.R., Groy A., Moss K.G., Minthorn E.A., Kulkarni S.G., Rominger C.M., Erskine S., Fisher K.E., Yang J. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- Gupta S., Ramjaun A.R., Haiko P., Wang Y., Warne P.H., Nicke B., Nye E., Stamp G., Alitalo K., Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Halilovic E., She Q.B., Ye Q., Pagliarini R., Sellers W.R., Solit D.B., Rosen N. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res. 2010;70:6804–6814. doi: 10.1158/0008-5472.CAN-10-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley M.E., Patrick D.M., Garcia M.R., Richardson J.A., Bassel-Duby R., van Rooij E., Olson E.N. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M.C., Maier C.R., Hobbs E.A., Ashmore A.R., Linnoila R.I., Dennis P.A. Akt1 deletion prevents lung tumorigenesis by mutant K-ras. Oncogene. 2011;30:1812–1821. doi: 10.1038/onc.2010.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E.L., Olive K.P., Tuveson D.A., Bronson R., Crowley D., Brown M., Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Ji H., Houghton A.M., Mariani T.J., Perera S., Kim C.B., Padera R., Tonon G., McNamara K., Marconcini L.A., Hezel A. K-ras activation generates an inflammatory response in lung tumors. Oncogene. 2006;25:2105–2112. doi: 10.1038/sj.onc.1209237. [DOI] [PubMed] [Google Scholar]
- Johnson L., Mercer K., Greenbaum D., Bronson R.T., Crowley D., Tuveson D.A., Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kim C.F., Jackson E.L., Kirsch D.G., Grimm J., Shaw A.T., Lane K., Kissil J., Olive K.P., Sweet-Cordero A., Weissleder R., Jacks T. Mouse models of human non-small-cell lung cancer: raising the bar. Cold Spring Harb. Symp. Quant. Biol. 2005;70:241–250. doi: 10.1101/sqb.2005.70.037. [DOI] [PubMed] [Google Scholar]
- Knight Z.A., Gonzalez B., Feldman M.E., Zunder E.R., Goldenberg D.D., Williams O., Loewith R., Stokoe D., Balla A., Toth B. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.H., Counter C.M. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell. 2005;8:381–392. doi: 10.1016/j.ccr.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- Maurer T., Garrenton L.S., Oh A., Pitts K., Anderson D.J., Skelton N.J., Fauber B.P., Pan B., Malek S., Stokoe D. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Arcas M., Hancock D.C., Sheridan C., Kumar M.S., Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI 3-kinase in KRAS mutant lung cancer. Cancer Discov. 2013;3:548–563. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Supprian M., Rajewsky K. Vagaries of conditional gene targeting. Nat. Immunol. 2007;8:665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- Shaw A.T., Kim D.W., Nakagawa K., Seto T., Crinó L., Ahn M.J., De Pas T., Besse B., Solomon B.J., Blackhall F. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- She Q.B., Halilovic E., Ye Q., Zhen W., Shirasawa S., Sasazuki T., Solit D.B., Rosen N. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima F., Yoshikawa Y., Ye M., Araki M., Matsumoto S., Liao J., Hu L., Sugimoto T., Ijiri Y., Takeda A. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc. Natl. Acad. Sci. USA. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopasakis V.R., Liu P., Suzuki R., Kondo T., Winnay J., Tran T.T., Asano T., Smyth G., Sajan M.P., Farese R.V. Specific roles of the p110alpha isoform of phosphatidylinsositol 3-kinase in hepatic insulin signaling and metabolic regulation. Cell Metab. 2010;11:220–230. doi: 10.1016/j.cmet.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sos M.L., Fischer S., Ullrich R., Peifer M., Heuckmann J.M., Koker M., Heynck S., Stückrath I., Weiss J., Fischer F. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc. Natl. Acad. Sci. USA. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Burke J.P., Phan J., Burns M.C., Olejniczak E.T., Waterson A.G., Lee T., Rossanese O.W., Fesik S.W. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Ogata H., Nishigaki R., Broide D.H., Karin M. Tobacco smoke promotes lung tumorigenesis by triggering IKKbeta- and JNK1-dependent inflammation. Cancer Cell. 2010;17:89–97. doi: 10.1016/j.ccr.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B., Zhao X., Meyerson M. Somatic alterations in the human cancer genome. Cancer Cell. 2004;6:433–438. doi: 10.1016/j.ccr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Zhao J.J., Cheng H., Jia S., Wang L., Gjoerup O.V., Mikami A., Roberts T.M. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc. Natl. Acad. Sci. USA. 2006;103:16296–16300. doi: 10.1073/pnas.0607899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.