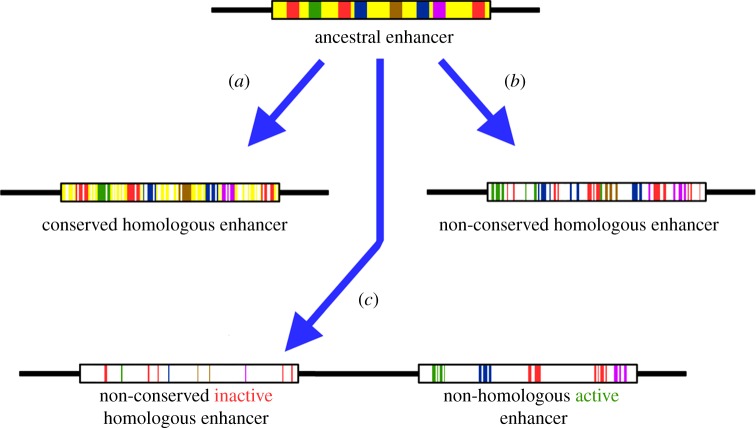
Figure 1.
Scheme of possible evolutionary paths of enhancers. Top: ancestral enhancer (box) has a given arrangement of transcription factor binding sites (TFBSs, coloured squares) in a background of near-neutral sequence (yellow). (a) As evolutionary time goes by, the enhancer can accumulate relatively few mutations (indicated as white areas in box) and give rise to homologous enhancers that can be aligned (i.e. are ‘conserved’) between species. (b) If TFBSs undergo much turnover and the accumulation of mutations is too high, homologous enhancers may no longer be alignable (i.e. be ‘non-conserved’). (c) The ancestral enhancer becomes inactive by the accumulation of mutations, but this is compensated by the appearance of a new enhancer elsewhere in the locus that fulfils the same regulatory function. In the path to (c), both functional enhancers coexist for some time until one of them becomes non-functional.