Fig. 3.
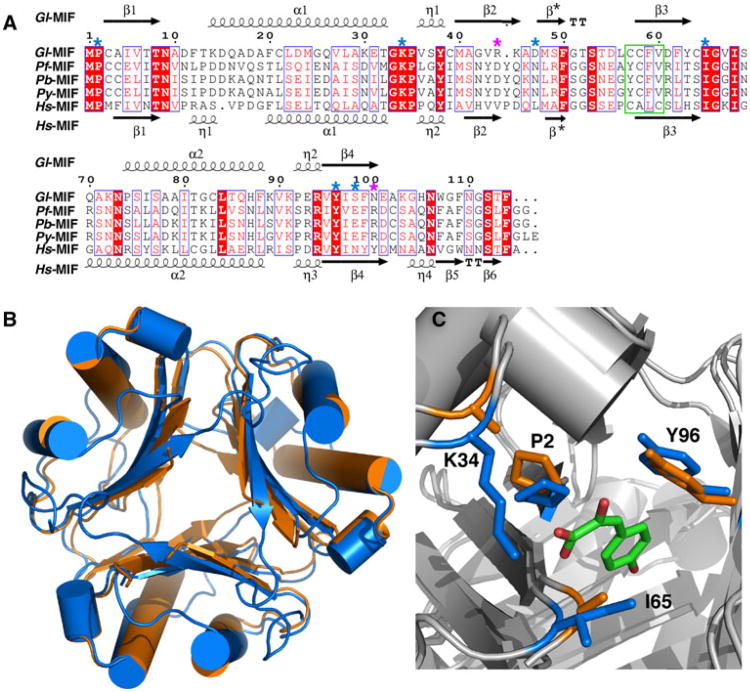
Sequence alignment of selected MIF proteins and structural comparison. a ClustalW2 amino acid sequence alignment of MIF proteins from G lamblia, P. falciparum, P. berghei, P. yoelii, and H. sapiens illustrated with ESPript [55]. Red shaded regions highlight invariant residues in the alignment with invariant and conserved residues inside a blue box. Key, evolutionarily conserved residues necessary for tautomerase activity are indicated by a blue asterisk above the Gl-MIF sequence. Position of the two gate-keeper residues is indicated with a purple asterisk above the Gl-MIF sequence. The oxidoreductase motif in Hs-MIF in outlined with a green box. b Superposition of the ribbon representations of the crystal structures of the Gl-MIF (3T5S, orange) and Hs-MIF (1MIF, blue) trimers. c Superposition of the crystal structures of Gl-MIF (3T5S) and Hs-MIF bound to p-hydroxyphenylpyruvate (HPP) (1CA7) highlighting the side chains of four of the important substrate binding residues (P2, K34, I65, Y96 in Gl-MIF) of the tautomerase site. The HPP is coloured by atom type (green = carbon, red = oxygen) and the Gl-MIF and Hs-MIF residues coloured orange and blue, respectively. The structure were superimposed using the program SuperPose [56]