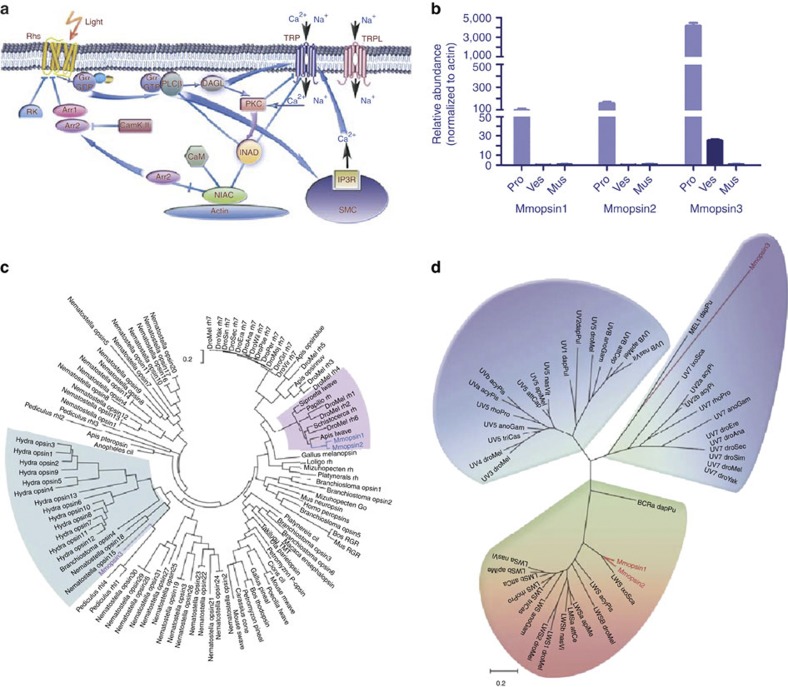
Figure 4. Molecular basis for photosensor function in the scorpion tail.
(a) Pathway of light-sensing signal transduction in the scorpion tail. The signal transduction network is modelled after that of D. melanogaster55. The involved signalling molecules are listed in Supplementary Tables S17 and S18. (b) Quantitative expression analysis of opsin genes, Mmopsin1, Mmopsin2 and Mmopsin3, in M. martensii. pro, prosoma; ves, vesicle or venom gland; mus, muscle from chelas and metasomal segments I-V. Data are expressed as the mean±s.d. from three replicates. (c) Phylogenetic relationships among opsins. The phylogenetic tree is constructed with the conserved sequences among opsin proteins, using ML method (MEGA5)56 (Supplementary Note 5). Mmopsin1 and Mmopsin2 are grouped with those from insects, whereas Mmopsin3 is closely related to opsins from genera Hydra, Branchiostoma and Nematostella. (d) Spectrum bias of the M. martensii opsins. The phylogenetic tree is constructed as in c. Mmopsin3 is a member of short-wavelength (ultraviolet to blue) opsins, but Mmopsin1 and Mmopsin2 belong to long-wavelength opsins.