Abstract
Fatty acid desaturases play critical roles in regulating the biosynthesis of unsaturated fatty acids in all biological kingdoms. As opposed to plants, mammals are so far characterized by the absence of desaturases introducing additional double bonds at the methyl-end site of fatty acids. However, the function of the mammalian fatty acid desaturase 3 (FADS3) gene remains unknown. This gene is located within the FADS cluster and presents a high nucleotide sequence homology with FADS1 (Δ5-desaturase) and FADS2 (Δ6-desaturase). Here, we show that rat FADS3 displays no common Δ5-, Δ6- or Δ9-desaturase activity but is able to catalyze the unexpected Δ13-desaturation of trans-vaccenate. Although there is no standard for complete conclusive identification, structural characterization strongly suggests that the Δ11,13-conjugated linoleic acid (CLA) produced by FADS3 from trans-vaccenate is the trans11,cis13-CLA isomer. In rat hepatocytes, knockdown of FADS3 expression specifically reduces trans-vaccenate Δ13-desaturation. Evidence is presented that FADS3 is the first “methyl-end” fatty acid desaturase functionally characterized in mammals.
Keywords: fatty acid desaturase; FADS cluster; desaturase activity; trans-vaccenic acid; conjugated linoleic acid; trans11,cis13-octadecadienoic acid; rumenic acid
FA desaturases introduce double bonds between defined carbons of the fatty acyl chain and catalyze crucial rate-limiting steps in the biosynthesis of unsaturated FAs. Desaturation activities are conserved across biological kingdoms, but the positioning of the inserted double bonds differs between angiosperm plants and higher animals (1). While plants produce linoleic acid (18:2 n-6) and α-linolenic acid (18:3 n-3) from oleic acid (18:1 n-9) using Δ12- and Δ15-desaturases (2), mammals typically lack these “methyl-end” FA desaturases. Therefore, linoleic acid and α-linolenic acid must be provided by the diet (3). These dietary essential FAs then serve as precursors for longer PUFAs of the n-6 and n-3 series, including arachidonic acid (20:4 n-6) and docosahexaenoic acid (22:6 n-3) required for many important physiological functions in humans (4, 5). In this PUFA biosynthesis, only “front-end” desaturases (Δ5- and Δ6-desaturases) are involved to introduce new double bonds between the preexisting double bonds and the carboxyl end of FAs (6).
In 2000, the genomic structure of the fatty acid desaturase (FADS) cluster, located on chromosome 11 in humans, was reported (7). This cluster includes the FADS1 and FADS2 genes that code, respectively, for the well-known Δ5- and Δ6-desaturases involved in PUFA biosynthesis (8–10). A third gene was identified on this cluster with high degree of nucleotide sequence homology with both FADS1 (62%) and FADS2 (70%) and was therefore named FADS3. Since its first description in humans (7), FADS3 has been identified in rats (11), baboons (12), mice (3), and many other mammals showing a similar nucleotide sequence and intron/exon organization (12). When analyzing the peptide sequence deduced from the FADS3 nucleotide sequence, FADS3 protein was identified as a putative desaturase, according to its similarity with FADS1 and FADS2. Indeed, the predicted structure of FADS3 is composed of an N-terminal cytochrome b5-like domain characterized by a HPGG motif shown to be essential for the Δ6-desaturase activity of FADS2 (13) and a C-terminal FA desaturase domain containing three histidine boxes (186HDLGH, 223HFQHH, and 387QIEHH in the rat) characteristic of front-end-desaturases (14). According to its amino acid sequence, FADS3 was thereafter supposed to be a mammalian membrane-bound front-end desaturase. However, no function has been attributed to the FADS3 protein yet (15).
In the absence of any described functional role, investigations have been focused on the tissue distribution of the FADS3 mRNA transcripts in different species. FADS3 mRNA was shown to be ubiquitously expressed in a large variety of tissues in humans (7), baboons (12), and rats (11), with a specific distribution compared with FADS1 and FADS2 mRNAs. In neonate baboon tissues, seven alternative transcripts of FADS3 were described (12), but when they were cloned and expressed into yeast and mammalian cells, no activity toward a variety of PUFA substrates was detected (16). Interestingly, the expression of the FADS3 transcripts was upregulated by dietary long-chain PUFAs, whereas FADS1 and FADS2 mRNA levels were decreased (17). In addition, gene polymorphism studies carried out in humans showed that several single nucleotide polymorphisms (SNP) located within the FADS3 gene were associated with variations in triglycerides, HDL- and LDL-cholesterol plasma levels (18, 19), and with variations in plasma PUFA levels (20–23). These correlations underlined the possible implication of FADS3 in lipid and FA homeostasis.
More studies were required to elucidate the function of the FADS3 protein. Here we have identified FADS3 as a trans-vaccenate (trans11-18:1) Δ13-desaturase. FADS3 is therefore the first methyl-end fatty acid desaturase functionally characterized in mammals. This discovery suggests that FADS3 may be an exception to the consensus dogma that rodents and higher animals are devoid of desaturases inserting double bonds in the methyl-end of fatty acids.
EXPERIMENTAL PROCEDURES
Plasmid constructs for expressing rat FADS1, FADS2, and FADS3 in COS-7 cells
The plasmids constructed for expressing rat FADS2 (pCMV/FADS2) and FADS3 (pCMV/FADS3) in mammalian cells have been described (11, 24). A plasmid coding for rat FADS1 was also constructed. Briefly, from the published (25) rat FADS1 cDNA sequence (GenBank accession number AF320509), oligonucleotide primers were designed to PCR amplify the entire coding sequence from the rat liver marathon-ready cDNA library (BD Biosciences). The forward primer (5′-gactccatggctcccgacccggtgcaga-3′) included the translation start codon (in bold) and NcoI restriction site (underlined). The reverse primer (5′-ctaggcggccgctattggtgaaggtaagcatccagccaga-3′) contained the translation stop codon (in bold) and NotI restriction site (underlined). The resulting PCR product was cloned into the pCMV expression vector using NcoI and NotI sites. The integrity of the plasmid, now referred to as pCMV/FADS1, was confirmed by DNA sequencing.
Rat FADS proteins were expressed by transiently transforming COS-7 cells with pCMV/FADS1, pCMV/FADS2, and pCMV/FADS3. Cells transfected with the pCMV empty were used as negative control. COS-7 cells were routinely maintained in DMEM containing 10% FCS (v/v), 50,000 IU/l penicillin, 50 mg/l streptomycin, and 10 mg/l gentamycin. The cells were seeded one day before transfection at 50% confluence and transfected the following day with 10 µg of plasmid for 1.5 × 106 cells by using the PolyFect reagent (Qiagen).
Extinction of native FADS3 and expression of recombinant FADS3 in cultured rat hepatocytes
The experimental procedure was approved by the French Animal Care Committee in compliance with recommendations of the 2013-118 directive for animal experimentation. The agreement reference of the laboratory to realize rat liver perfusion is R-2012-FP-01. Hepatocytes were obtained from male rats after an in situ collagenase perfusion of the liver, as described (26). Small interfering RNA (siRNA) was obtained from Sigma. Negative siRNA control targets unrelated mRNA sequences, and siRNA against FADS3 targets 5′-gcgacaaguggcuggucau-3′ sequence. After the rat liver perfusion, hepatocytes were seeded at 2.0 × 106 cells in 6 cm diameter culture dishes. They were transfected at the same time with 25 nM of either siRNA control or siRNA targeting FADS3 by using the N-TER Nanoparticle siRNA transfection system (Sigma). For the expression of recombinant FADS3, hepatocytes were transfected after 4 h of culture with 10 µg of plasmid/1.6 × 106 cells by using the jetPEI-Hepatocyte kit (Polyplus-transfection).
Incubation of cells with fatty acids
The functionality of the FADS proteins was investigated by incubating the transfected COS-7 cells or cultured rat hepatocytes with different FA. The common Fas, including trans-vaccenic acid (TVA), were obtained from Sigma (Saint-Quentin-Fallavier, France). Rumenic acid (cis9,trans11-18:2 or RA) and a mixture of cis11,trans13-18:2, cis11,cis13-18:2, and trans11,trans13-18:2 were from Matreya (Pleasant Gap, PA). The pure trans11,trans13-18:2 was obtained from Larodan (Malmö, Sweden). [1-13C]trans-vaccenic acid (chemical purity 95%, isotopic enrichment 99%) was synthesized by the Commissariat à l'Energie Atomique (CEA, Gif-sur-Yvette, France) according to well-established procedures (27, 28). FA albuminic complexes were prepared as described (26). The final FA concentration of the incubation media was 200 µM and albumin concentration was 150 µM, leading to a FA:albumin molar ration of 1.3:1. At 24 h post transfection (COS-7 cells) or 24 h after plating (hepatocytes), the incubation was initiated by replacing the culture medium with the FA-containing medium. Incubation was then carried out for 24 h at 37°C in a 5% CO2 atmosphere.
Fatty acid analysis
Cellular lipids were extracted with hexane-isopropanol (3:2, v/v) after acidification with 1 ml HCl 3 M, as described (26). After saponification for 15 min at 70°C with 1 ml of 0.5 M NaOH in methanol, FAs were methylated with BF3 (14% in methanol) at 70°C for 10 min. The fatty acid methyl esters (FAME) were extracted with pentane and analyzed by GC-MS using an Agilent Technologies 7890N (Bios Analytic) with a bonded silica capillary column (60 m × 0.25 mm; BPX 70; SGE). Helium was used as carrier gas (average velocity 24 cm/s). The column temperature program started at 150°C, ramped at 2°C/min to 220°C, and held at 220°C for 10 min. Mass spectra were recorded with an Agilent Technologies 5975C inert MSD with triple-axis detector. The mass spectrometer was operated under electron impact ionization conditions (electron energy 70 eV, source temperature 230°C). Data were obtained in scan mode with a mass range of m/z 50–550 atomic mass unit (amu). Identification of FAMEs was based upon retention times (Rt) obtained for methyl ester of authentic standards, when available. The National Institute of Standards and Technology (NIST) database (version 2.01) was sometimes used to identify unknown FAs.
For positional analysis of carbon double bonds, FAMEs were converted to 4,4-dimethyloxazoline (DMOX) derivatives (29, 30) and analyzed by GC-MS in electron impact ionization mode. The column, gas vector, and detector were similar to those used for the analysis of FAMEs (see above). To preconcentrate the minor FA from some experiments before the DMOX derivatization, FAMEs were separated according to the number and configuration of their double bonds on preparative silver ion thin-layer chromatography using silica gel H impregnated with 10% (w/w) AgNO3 and a mixture of 90/10 (v/v) hexane/diethylether for development (31).
Immunoblot analysis
Immunoblotting was performed as described (11) using the antibodies described here. Rat FADS1 protein was detected with a polyclonal antibody to FADS1 targeting the antigenic peptide 100QPSFEPTKNKALTDE. Rat FADS2 was detected with an antibody to FADS2 directed against the peptide 3KGGNQGEGSTEL. Rat FADS3 was detected with an antibody to FADS3 (11) directed against the peptide 352PKEIGHEKHRDWAS. An antibody to stearoyl-CoA desaturase 1 (SCD1) was obtained from Santa Cruz Biotechnology. A quantitative estimation of protein loading was performed with an antibody to actin (Sigma).
RNA isolation and quantitative RT-PCR
Total RNA was extracted from hepatocytes with Trizol (Invitrogen). RNA was converted into cDNA with the IScript cDNA synthesis kit (Bio-Rad). RNA was quantified by real-time PCR with the Taqman Universal PCR Master Mix (Applied Biosystems) containing 40 ng of retrotranscripted RNA, 0.5 µM of primers, and 0.25 µM of Taqman probe. Primers and 5′-FAM/TAMRA-3′ Taqman probes specific to each rat FADS gene were described previously (11). Primers (forward primer 5′-caaagagaagggcggaaagc-3′ reverse primer 5′-aaagtttcgccccagcagta-3′) and a Taqman probe (5′FAM-ctggcctcctgctcatgtgcttca-TAMRA3′) were also designed to quantify SCD1 mRNA. Amplification was performed by an ABI PRISM 7000 (Applied Biosystems) over 40 cycles of 95°C for 15 s and 60°C for 1 min. The 18S gene expression was quantified as an endogenous control using the 18S RNA Control kit Yakima-Yellow Eclipse Dark Quencher (Eurogentec), and relative gene expression was determined from the cycle thresholds (Ct) using the ΔΔCt method.
Expression of results and statistical analysis
All data values shown represent the mean ± SD for the number of replicates (n) indicated in each figure legend. Student t-test was used to determine the statistical significance between groups, where indicated.
RESULTS
Rat FADS3 is not functionally redundant with FADS1 or FADS2
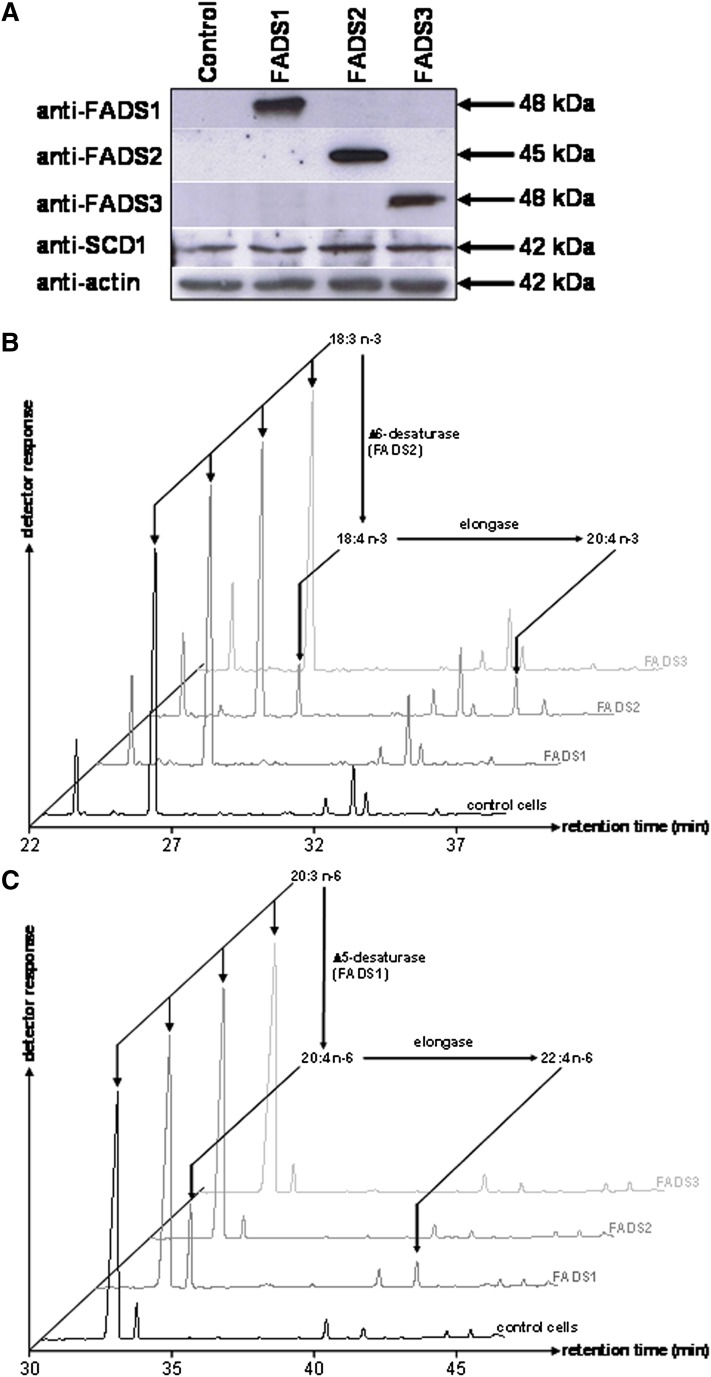
To investigate the putative desaturase activity of FADS3, COS-7 cells expressing rat FADS1, FADS2, or FADS3 were grown for 24 h in culture media supplemented with various potential FA substrates. Immunoblot analyses (Fig. 1A) revealed that transient transfection resulted in the specific expression of each FADS protein. Rat FADS1 and FADS2 were detected as single immunoreactive bands with respective apparent molecular weights of 48 and 45 kDa. As previously described (11), recombinant rat FADS3 was specifically detected at about 48 kDa. Endogenous FADS1, FADS2, and FADS3 were not detected in control cells with our anti-rat antibodies, but COS-7 cells endogenously expressed the Δ9-desaturase (SCD1).
Fig. 1.
Expression and desaturase activities of recombinant rat FADS1, FADS2, and FADS3 in COS-7 cells. (A) COS-7 cells were transfected with the following plasmids: pCMV empty (control cells), pCMV/FADS1, pCMV/FADS2, and pCMV/FADS3. Immunoblots were assessed on COS-7 cell extracts transiently expressing the recombinant FADS proteins. Recombinant FADS1, FADS2, and FADS3 were detected with polyclonal antibodies targeting specific antigenic peptides of these proteins. Quantitative estimation of protein loading was performed with an anti-actin antibody. Molecular weights are indicated in kDa. (B) GC-MS profiles of FAMEs extracted from COS-7 cells expressing rat FADS1, FADS2, or FADS3 and incubated with α-linolenic acid (18:3 n-3) (C) GC-MS profiles of FAMEs extracted from COS-7 cells expressing rat FADS1, FADS2, or FADS3 and incubated with eicosatrienoic acid (20:3 n-6). Control COS-7 cells and COS-7 cells transiently expressing the recombinant FADS proteins were incubated for 24 h with albumin-bound FA (200 µM). The identity of each important FA is indicated above its respective peak. Representative example of n = 5 distinct experiments.
After 24 h of incubation with a FA, the FAME profile of cells expressing each FADS protein was analyzed by GC-MS to detect the presence of newly desaturated FA. Cells expressing rat FADS2 were able to synthesize 18:4 n-3 from 18:3 n-3 (Fig. 1B), whereas control cells and cells expressing FADS1 or FADS3 were unable to Δ6-desaturate 18:3 n-3. In our experimental conditions, recombinant FADS2, but not FADS1 or FADS3, was also able to convert 18:2 n-6 into 18:3 n-6, 18:1 n-9 into 18:2 n-9, and 16:1 n-7 into 16:2 n-7 (data not shown). Moreover, only the cells expressing rat FADS1 were able to produce higher amounts of 20:4 n-6 from 20:3 n-6 (Fig. 1C). These results validated that our recombinant model was consistent with the known in vivo Δ5- and Δ6-desaturase activities and FA substrates of FADS1 and FADS2 (24, 29) and showed that FADS3 was not functionally redundant with FADS1 and FADS2. Similar experiments carried out with saturated FA (12:0, 14:0, 16:0, and 18:0) also showed that FADS3 displayed no Δ9-desaturase activity (data not shown).
Rat FADS3 converts trans-vaccenate into an unknown 18:2
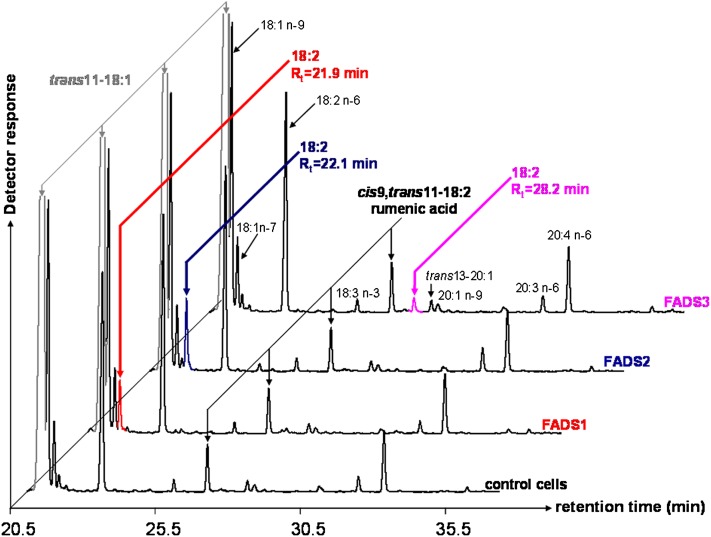
By using COS-7 cells expressing each rat FADS protein, many FA were unsuccessfully tested as putative desaturation substrates of FADS3. When TVA (trans11-18:1) was used, interesting FAME profiles were obtained (Fig. 2). First, whatever the experimental condition (control or transfected cells), TVA was converted to RA (cis9,trans11-18:2) by the native Δ9-desaturase (SCD1) present in COS-7 cells (Fig. 1A). RA (2.4 ± 0.6% of FA, n = 5) was identified with its retention time (Rt = 27.5 min) and MS profile after conversion to DMOX derivatives (see below). Second, in the FAME profile obtained with cells expressing FADS3, an additional specific FAME (with Rt = 28.2 min) was shown (Fig. 2), which was absent in control cells and in cells expressing FADS1 or FADS2. This FA (0.6 ± 0.2% of FA, n = 5) was identified as an octadecadienoic acid (18:2) with a FAME molecular ion at m/z 294 amu. It was in the range of Rt in which conjugated linoleic acids (CLA) are detected. Third, in the FAME profiles obtained with cells expressing either rat FADS1 or FADS2 (Fig. 2), additional octadecadienoic acids (18:2) with molecular ions at m/z 294 amu were detected with Rt = 21.9 min and Rt = 22.1 min, respectively.
Fig. 2.
GC-MS profiles of FAMEs extracted from COS-7 cells expressing rat FADS1, FADS2, or FADS3 and incubated with TVA. Control COS-7 cells and COS-7 cells transiently expressing the recombinant FADS proteins were cultivated for 24 h with albumin-bound TVA (200 µM). The identity of each important FA is indicated above its respective peak. Rumenic acid (Rt = 27.5 min) was detected in all the experimental conditions (control cells and transfected cells). Three different and unknown octadecadienoic acids (with FAME molecular ions at m/z 294) were produced in COS-7 cells expressing FADS1 (Rt = 21.9 min), FADS2 (Rt = 22.1 min,) and FADS3 (Rt = 28.2 min). Representative example of n = 5 distinct experiments.
Thereafter, similar experiments were carried out with [1-13C]TVA. The obtained FAME profiles were identical to those described in Fig. 2, and they confirmed the presence of the three unknown 13C-18:2, with FAME molecular ions at m/z 295 amu, when recombinant FADS3, FADS2, and FADS1 were expressed.
Rat FADS3 is a trans-vaccenate desaturase, not a rumenate isomerase
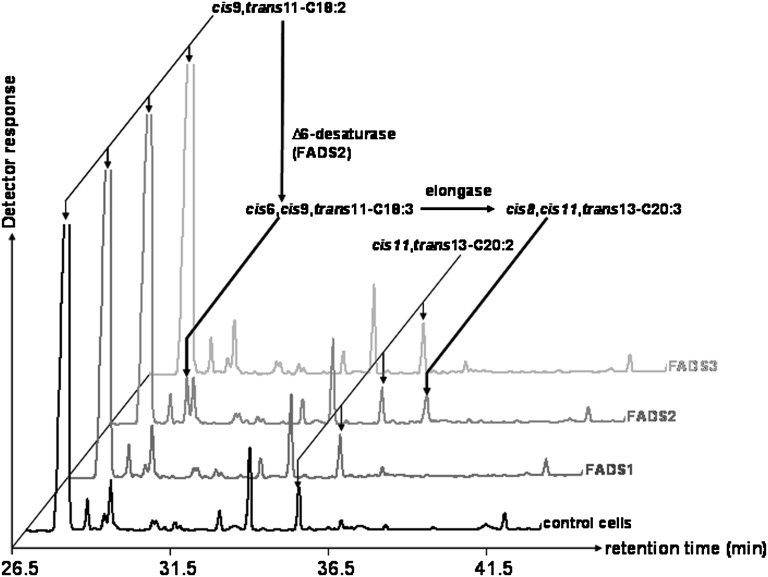
The results described above showed that COS-7 cells expressing FADS3 and incubated with TVA gained the ability to produce an unknown 18:2. However, at this stage, we were not able to discriminate between a desaturase activity of the recombinant FADS3 toward TVA and an isomerase activity toward RA, which was concomitantly produced from TVA by the endogenous Δ9-desaturase (Fig. 2). Therefore, RA was subsequently incubated with COS-7 cells expressing rat FADS1, FADS2, or FADS3. In the FAME profile obtained after RA incubation with cells expressing FADS3 (Fig. 3), no additional peak with Rt = 28.2 min was detected compared with control cells. This result indicated that, in our experimental conditions, the recombinant FADS3 was not able to catalyze the isomerization of RA and suggested that the 18:2 shown in FADS3-expressing cells (Fig. 2) was likely to result from TVA desaturation.
Fig. 3.
GC-MS profiles of cellular FAMEs extracted from COS-7 cells expressing rat FADS1, FADS2, or FADS3 and incubated with rumenic acid (cis9,trans11-CLA). Control COS-7 cells and COS-7 cells transiently expressing the recombinant FADS proteins were cultivated for 24 h with albumin-bound rumenic acid (200 µM). The identity of each important FA is indicated above its respective peak. It shows that only the cells expressing FADS2 were able to use RA as a substrate. Indeed, detectable amounts of an octadecatrienoic acid (18:3) with FAME molecular ion at m/z 292 amu and Rt = 29.1 min were produced when compared with control cells. Although not fully characterized in the present study, this FA was identified as the cis6,cis9,trans11-18:3, the known Δ6-desaturated product of RA (56–58), by using the NIST database. Representative example of n = 3 distinct experiments.
Structural characterization strongly suggests that the 18:2 produced by rat FADS3 is the trans11,cis13-CLA isomer
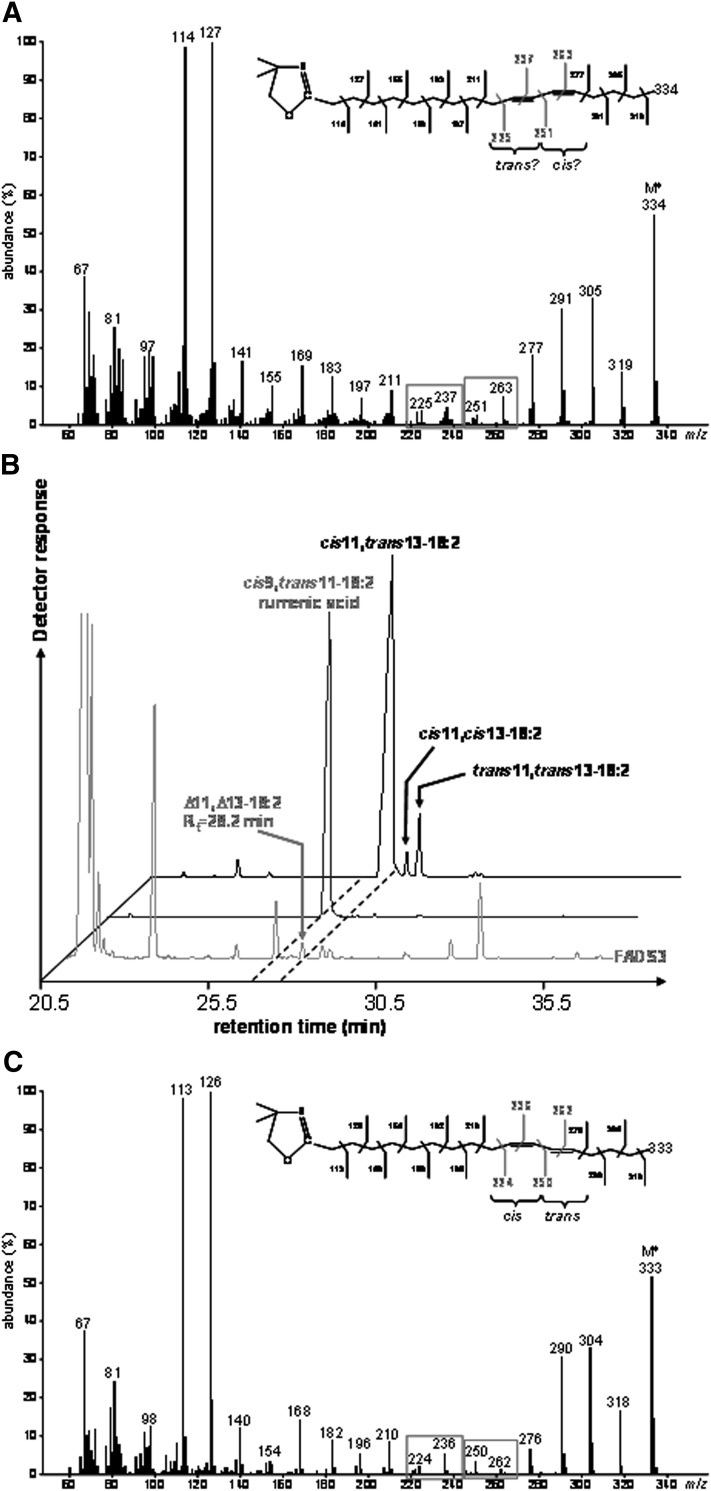
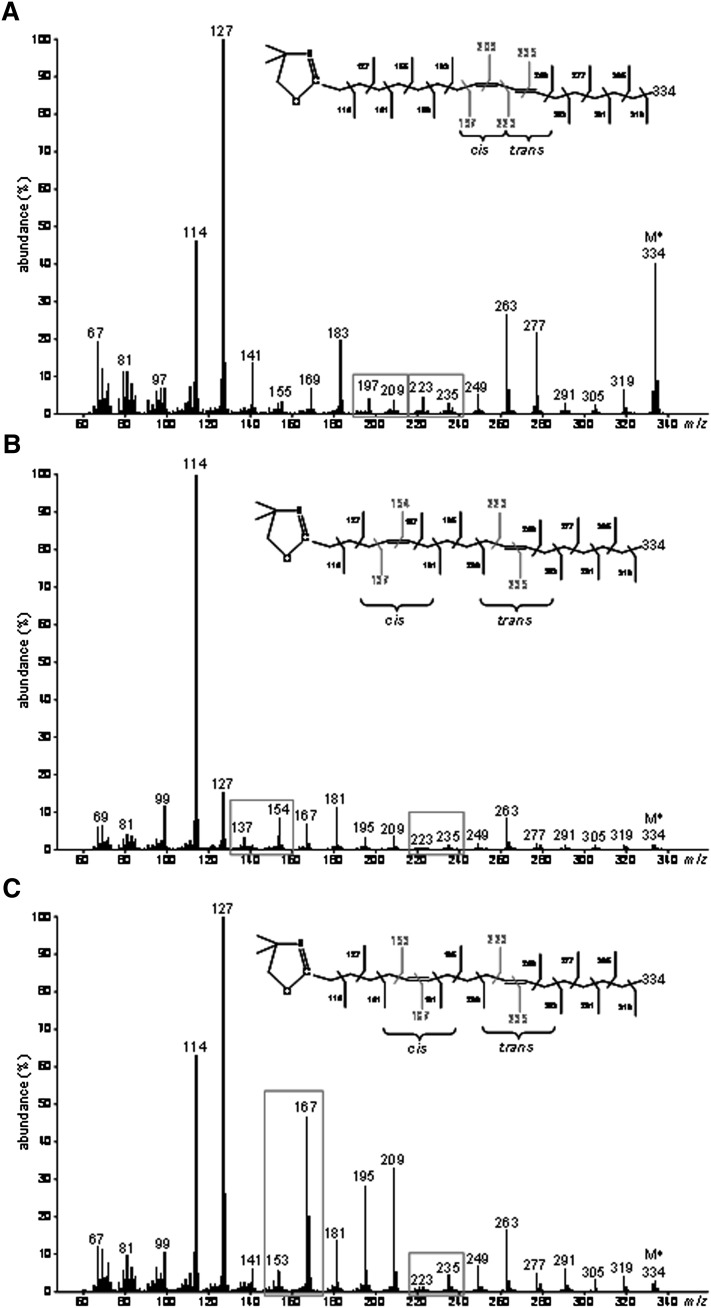
To locate the double bonds in the carbon chain of the unknown 18:2 produced from TVA in cells expressing FADS3, cellular FA obtained after incubation with [1-13C]TVA were converted to DMOX derivatives and analyzed by GC-MS. The mass spectrum of the unknown 13C-18:2 derivative (Fig. 4A) contained DMOX-characteristic intense ions at m/z 114 and 127 amu and a molecular ion at m/z 334 amu, which confirmed that it possessed a di-unsaturated 13C18 structure. A gap of 12 amu was detected between ions containing 10 and 11 carbons (m/z 225 and 237 amu) and between ions containing 12 and 13 carbons (m/z 251 and 263 amu), showing that the unknown FA was a Δ11,13-CLA (Fig. 4A). Analysis of this derivative strongly suggested that, after its conversion from [1-13C]TVA, the 13C label was still positioned at the 1st carbon.
Fig. 4.
Identification of the unknown octadecadienoic acid produced from trans-vaccenate in cells expressing rat FADS3 as a Δ11,13-conjugated linoleic acid isomer. (A) Electron-impact mass spectrum of the DMOX derivative of the unknown 13C-octadecadienoic acid obtained after incubation of COS-7 cells expressing FADS3 with [1-13C]TVA. (B) GC-MS profile of the FAMEs extracted from COS-7 cells expressing FADS3 and incubated with TVA compared with commercial rumenic acid and a commercial mixture of cis11,trans13-18:2, cis11,cis13-18:2, and trans11,trans13-18:2 as FAME standards. (C) Electron-impact mass spectrum of the DMOX derivative of the commercial cis11,trans13-18:2.
To get more information on the geometrical structure of this Δ11,13-CLA, we used a commercial mixture of cis11,trans13-CLA, cis11,cis13-CLA, and trans11,trans13-CLA as FAME standards (the trans11,cis13-CLA isomer was not commercially available). GC-MS analysis of these standards (Fig. 4B) showed that the cis11,cis13 and trans11,trans13-CLA isomers had significantly higher retention times (Rt = 28.8 and 29.1 min, respectively) than the Δ11,13-CLA produced by FADS3 from TVA (Rt = 28.2 min), while the cis11,trans13-CLA isomer had a slightly lower retention time (Rt = 28.0 min). Knowing that the four geometrical isomers of a CLA elute with a specific order (cis-trans, trans-cis, cis-cis, and trans-trans) when separated on a capillary column with a polar stationary phase (32), this result first suggested that the unknown Δ11,13-CLA was not the cis11,cis13 or the trans11,trans13 isomer. However, because the difference in Rt between the cis11,trans13 standard isomer and the unknown Δ11,13-CLA was small (only 0.2 min), identification remained uncertain between the trans11,cis13 and the cis11,trans13 geometric isomers. Therefore, we compared the DMOX mass spectrum pattern of the unknown Δ11,13-CLA (Fig. 4A) with that of the cis11,trans13 standard isomer with the nearest Rt (Fig. 4C). We noticed that, in the case of a trans-Δ13 double bond, the ion at m/z 250 amu was slightly more abundant than the ion at m/z 262 amu, whereas the ion at m/z 251 amu of the unknown 13C-labeled Δ11,13-CLA was significantly less abundant than the ion at m/z 263 amu (Fig. 4A). Although there was no commercially available standard for comparison and complete conclusive identification, these observations strongly suggested that this FA was likely the trans11,cis13-CLA corresponding to a cis-Δ13-desaturated product from TVA in cells expressing FADS3. This result was in agreement with the Δ11 double bond preexisting in TVA keeping its trans configuration when the second cis-Δ13 double bond was inserted by the recombinant FADS3.
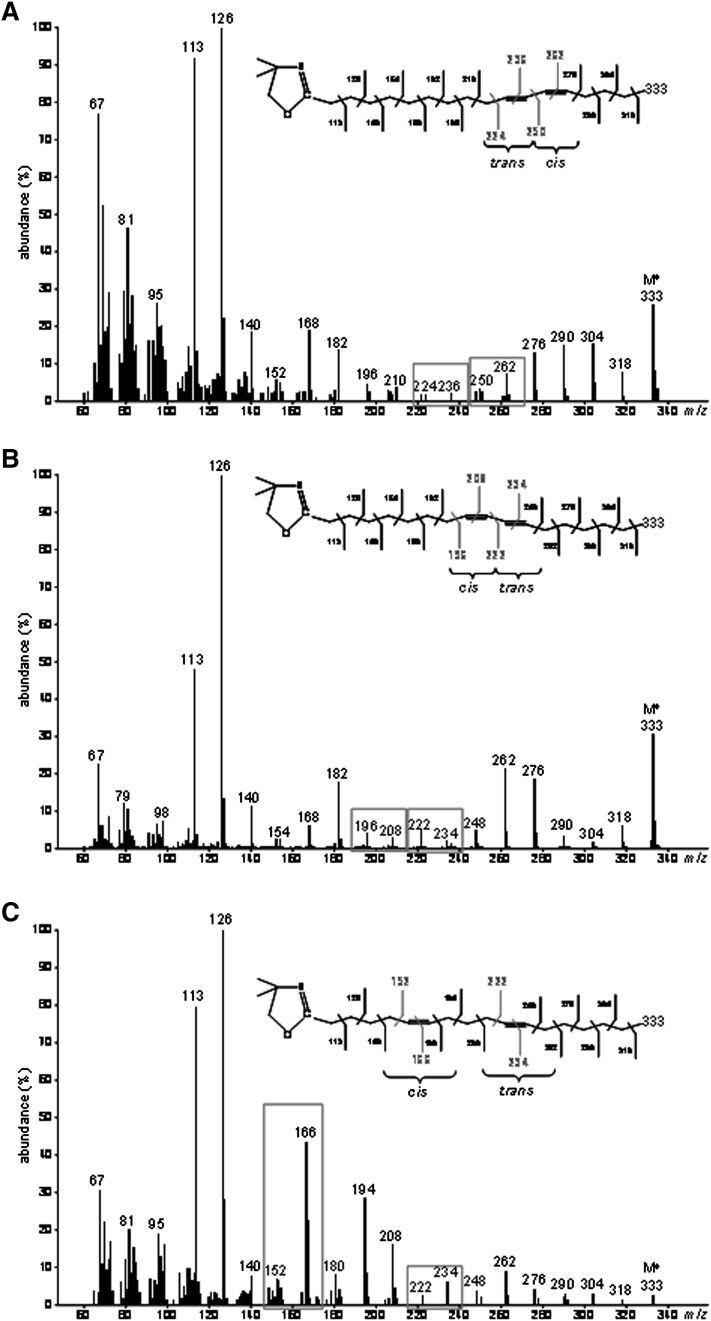
GC-MS analyses of DMOX derivatives also allowed the identification of 13C-cis9,trans11-18:2 (RA) produced by endogenous Δ9-desaturation of [1-13C]TVA (Fig. 5A), 13C-cis5,trans11-18:2 produced by recombinant FADS1 (Fig. 5B) and 13C-cis6,trans11-18:2 produced by recombinant FADS2 (Fig. 5C).
Fig. 5.
Electron-impact mass spectra of DMOX derivatives of the different 13C-octadecadienoic acids obtained after incubation of [1-13C]TVA with COS-7 cells expressing either rat FADS1 or FADS2. Molecular ions (M+) are m/z 334. (A) 13C-rumenic acid (Rt = 27.5 min) obtained after incubation of [1-13C]TVA with COS-7 cells in all the experimental conditions. (B) Unknown 13C-octadecadienoic acid (Rt = 21.9 min) obtained after incubation of [1-13C]TVA with COS-7 cells expressing rat FADS1. A gap of 12 amu was shown between ions containing 10 and 11 carbons (m/z 223 and 235 amu) locating this double bond in position 11-12, while the ion at m/z 154 amu was a diagnostic guide for a double bond in position 5-6 (59). This fatty acid was therefore identified as Δ5,11-18:2 (cis5,trans11-18:2) (C) Unknown 13C-octadecadienoic acid (Rt = 22.1 min) obtained after incubation of [1-13C]TVA with COS-7 cells expressing rat FADS2. A gap of 12 amu was shown between ions containing 10 and 11 carbons (m/z 223 and 235 amu) locating this double bond in position 11-12, while the fingerprint ions at m/z 153, 167, and 181 amu identified a double bond in position 6-7 (60). This fatty acid was therefore identified as Δ6,11-18:2 (cis6,trans11-18:2). Representative example of n= 3 distinct experiments.
FADS3 Δ13-desaturase activity is specific for trans-vaccenate
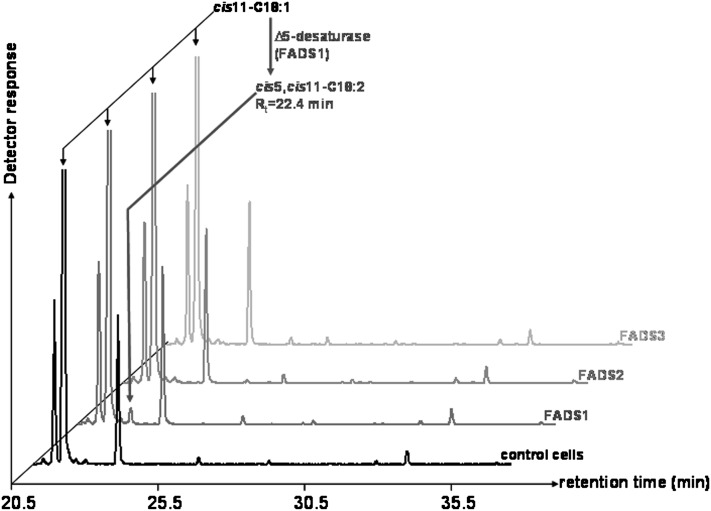
To investigate the substrate specificity of the newly discovered Δ13-desaturase activity of FADS3, several trans and cis monounsaturated FA were thereafter incubated with COS-7 cells expressing rat FADS3. In our experimental conditions, no desaturation of cis-vaccenate (cis11-18:1 or CVA) was shown in COS-7 cells expressing rat FADS3 (Fig. 6), as opposed to its trans isomer. Moreover, incubation of oleic and elaidic acids (cis9-18:1 and trans9-18:1), palmitoleic and palmitelaidic acids (cis9-16:1 and trans9-16:1), and cis11- and trans11-eicosenoic acids (cis11-20:1 and trans11-20:1) with COS-7 cells expressing rat FADS3 did not show any new desaturated FA (data not shown). Therefore, until now, TVA is the only FA that has been shown to be desaturated in the presence of recombinant rat FADS3.
Fig. 6.
GC-MS profile of cellular FAMEs extracted from COS-7 cells expressing rat FADS1, FADS2, or FADS3 and incubated with cis-vaccenate. Control COS-7 cells and COS-7 cells transiently expressing the recombinant FADS proteins were cultivated for 24 h with albumin-bound cis-vaccenate (200 µM). The identity of each important FA is indicated above its respective peak. It shows that only the cells expressing FADS1, but not FADS2 or FADS3, were able to desaturate CVA into an octadecadienoic acid (Rt = 22.4 min), which was identified as the cis5,cis11-18:2 by the NIST database. Representative example of n = 3 distinct experiments.
Biosynthesis of the suspected trans11,cis13-CLA in rat hepatocytes incubated with TVA
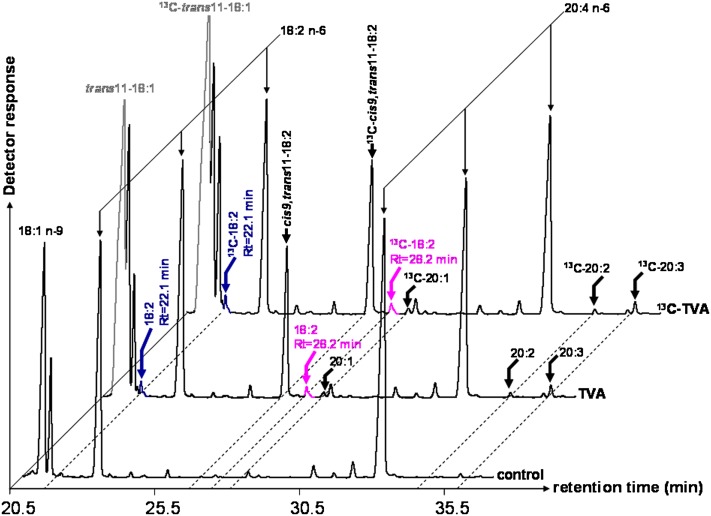
Because the liver is known to possess high FA desaturase activities (3), we chose cultured rat hepatocytes to investigate the ability of the native FADS3 to desaturate TVA, like its recombinant counterpart. Rat hepatocytes were incubated for 24 h with media supplemented with TVA or [1-13C]TVA and with a control medium without FA. RA (Rt = 27.5 min) produced by Δ9-desaturation of TVA was easily detected (7.6 ± 3.2% of FA, n = 5) in the cells incubated with TVA or 13C-TVA (Fig. 7). A small peak (0.4 ± 0.1% of FA, n = 5) with Rt = 28.2 min and FAME molecular ion at m/z 294 amu (incubation with 12C-TVA) or m/z 295 amu (incubation with 13C-TVA), similar to the suspected trans11,cis13-18:2 previously characterized in COS-7 cells expressing FADS3 (Fig. 2), was specifically detected (Fig. 7). The presence of another small 18:2 (0.4 ± 0.1% of FA, n = 5), with Rt = 22.1 min and FAME molecular ion at m/z 294 amu or m/z 295 amu (incubation with 12C-TVA or 13C-TVA), similar to the Δ6-desaturated product (cis6,trans11-18:2) of TVA by recombinant FADS2 in COS-7 cells (Fig. 2), was also shown (Fig. 7).
Fig. 7.
GC-MS profiles of FAMEs extracted from cultured rat hepatocytes incubated with TVA and [1-13C]TVA. Rat hepatocytes were incubated for 24 h with albumin-bound trans-vaccenate, [1-13C]trans-vaccenate, or no FA (control medium). The identity of each important FA is indicated above its respective peak. Representative example of n = 5 distinct experiments.
At this stage, analysis of DMOX derivatives (Fig. 8) was again performed to confirm the identity of the different 18:2 shown after incubation of TVA with cultured hepatocytes. The suspected trans11,cis13-18:2 (Fig. 8A), the cis9,trans11-18:2 (Fig. 8B) and cis6,trans11-18:2 (Fig. 8C) were clearly identified by their DMOX mass spectra, similar to those obtained in COS-7 cells (Figs. 4 and 5). Therefore, the Δ11,13-CLA isomer and cis6,trans11-18:2 already characterized in COS-7 cells expressing rat FADS3 and FADS2, respectively, were also found in rat hepatocytes incubated with TVA.
Fig. 8.
Electron-impact mass spectra of DMOX derivatives of the different octadecadienoic acids obtained after incubation of TVA with cultured rat hepatocytes. Molecular ions (M+) are m/z 333. (A) Octadecadienoic acid (Rt = 28.2 min) similar to the suspected trans11,cis13-C18:2 obtained in COS-7 cells expressing rat FADS3. (B) Rumenic acid (Rt = 27.5 min). (C) Octadecadienoic acid (Rt = 22.1 min) similar to the cis6,trans11-C18:2 obtained in COS-7 cells expressing rat FADS2.
Silencing of FADS3 expression in rat hepatocytes
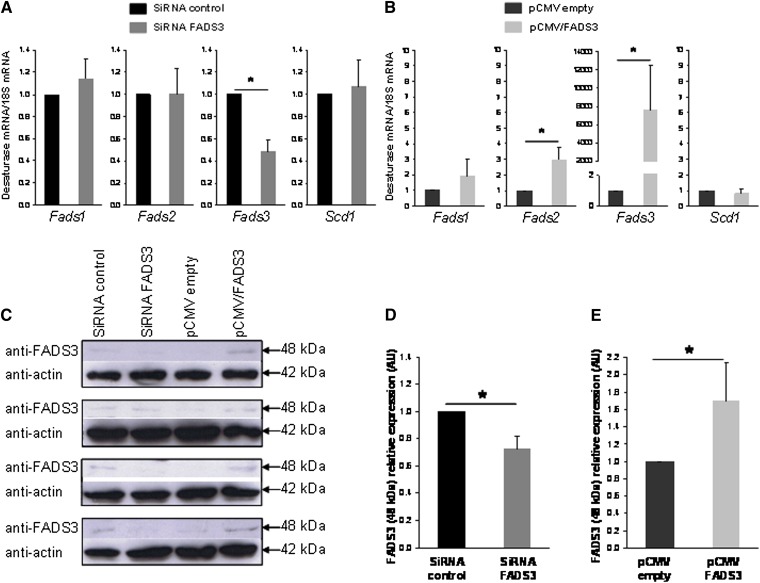
The synthesis of Δ11,13-18:2 from TVA described above suggested that native FADS3 was present and biologically active to catalyze this unique Δ13-desaturation in rat hepatocytes. To correlate the level of Δ11,13-18:2 synthesis with the level of FADS3 protein expression, hepatocytes were transfected with siRNA control or siRNA targeting FADS3 for 24 h and incubated for a further 24 h with TVA. Similarly, hepatocytes were transfected with the pCMV empty or pCMV/FADS3 for 24 h before incubation with TVA. Compared with siRNA control, FADS3 siRNA significantly suppressed the FADS3 mRNA expression by more than 50% (Fig. 9A). FADS1, FADS2, and SCD1 mRNA levels were not affected by the treatment. Conversely, pCMV/FADS3 transfection drastically increased the FADS3 mRNA expression (Fig. 9B). To a much lesser extent, FADS2 mRNA was increased in hepatocytes transfected with the pCMV/FADS3. In these experiments, because of the massive presence of FADS3 mRNA and high homology with FADS2 mRNA sequence, FADS2 mRNA may have been artifactually increased during RT-PCR analyses.
Fig. 9.
Effect of native FADS3 extinction and recombinant FADS3 expression on the levels of FADS3 mRNA detected by qRT-PCR and FADS3 protein detected by immunoblotting in culture rat hepatocytes. Hepatocytes were transfected with siRNA control or siRNA against Fads3 for 24 h and incubated for a further 24 h with TVA. Similarly, hepatocytes were transfected with the pCMV empty or pCMV/FADS3 for 24 h before incubation with TVA. (A, B) The cells were collected and analyzed for Fads1, Fads2, Fads3, and Scd1 mRNA expression by qRT-PCR in each experimental condition. (C) Expression of the 48 kDa immunoreactive band of FADS3 in rat hepatocytes assessed by immunoblot in each of the four distinct experiments realized. (D, E) Gray level quantification of the immunoreactive band at 48 kDa relative to actin in each experimental condition. Data are presented as mean ± SD (n = 4).
Although integral membrane-bound desaturases are notoriously difficult to study in native conditions (33), we also analyzed the expression of the FADS3 protein in each experimental condition (Fig. 9C). An immunoreactive band was hardly detected but reproducibly shown at 48 kDa with the anti-FADS3 antibody, matching the apparent molecular weight of the recombinant FADS3 (Fig. 1A). The intensity of the 48 kDa band was reduced approximately 30% in the cells treated with FADS3 siRNA compared with siRNA control (Fig. 9D) and increased by 1.7 in the cells transfected with pCMV/FADS3 compared with control hepatocytes (Fig. 9E).
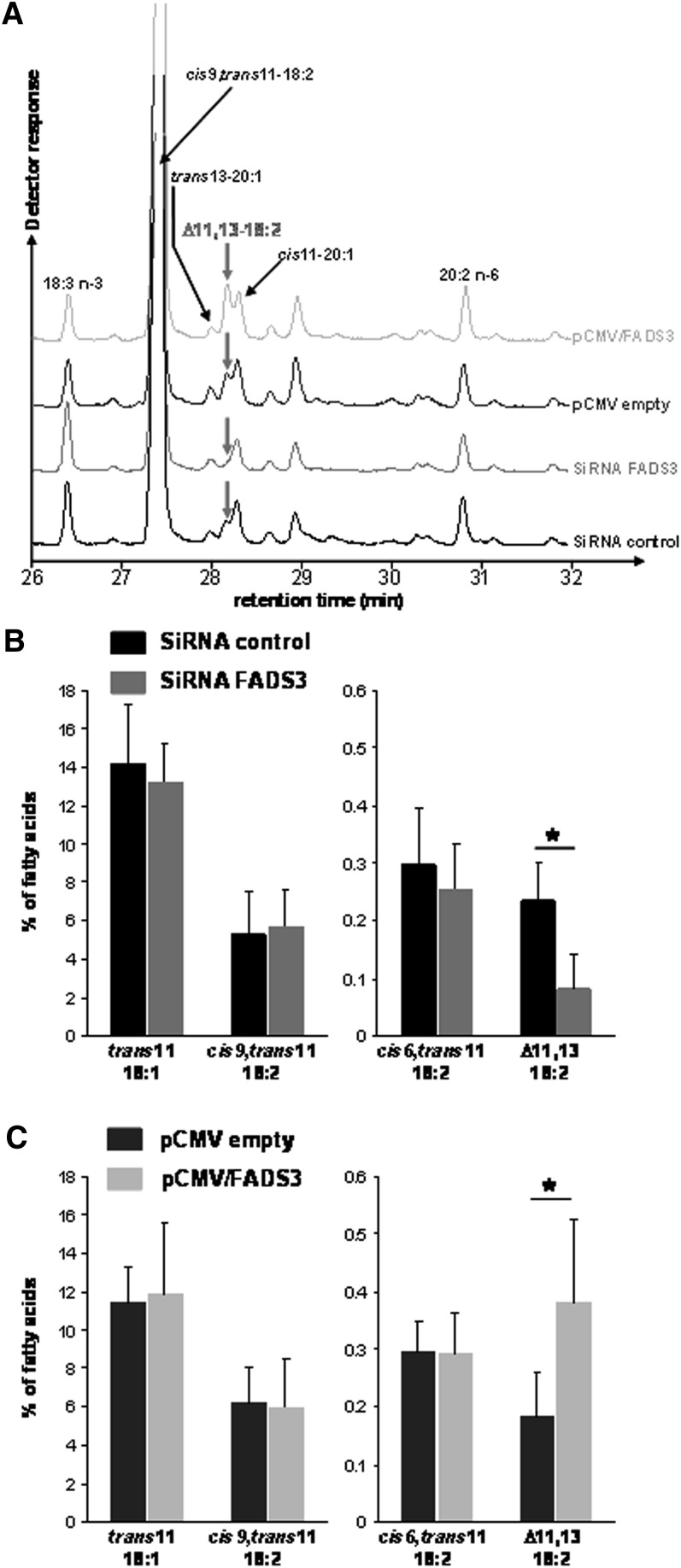
In the corresponding FAME profiles (Fig. 10), the peak corresponding to the suspected trans11,cis13-18:2 was 3-fold smaller in FADS3 siRNA transfected cells compared with siRNA control (Fig. 10A, B). Conversely, the Δ11,13-18:2 was 2-fold greater in the cells transfected with pCMV/FADS3 compared with the cells transfected with the pCMV empty (Fig. 10A, C). The amounts of TVA, RA, and cis6,trans11-18:2 were not affected by the treatments (Fig. 10B, C). In these experiments, due to the use of transfection reagents, TVA uptake and incorporation into the cells was lower than in classical incubation experiments (Fig. 7), leading therefore to a lower production of the Δ11,13-CLA.
Fig. 10.
Effect of native FADS3 extinction and recombinant FADS3 expression on the conversion of trans-vaccenate into the suspected trans11,cis13-CLA in rat hepatocytes. Hepatocytes were transfected with siRNA control or siRNA against Fads3 for 24 h and incubated for a further 24 h with TVA. Similarly, hepatocytes were transfected with the pCMV empty or pCMV/FADS3 for 24 h before incubation with TVA. (A) GC-MS profiles of FAMEs extracted from cultured rat hepatocytes in each experimental condition. For this set of experiments, the GC column, which was previously used for all the other GC-MS analyses, was changed, and isomers of eicosenoic acid (20:1) were eluted in the same region of the chromatogram as the suspected trans11,cis13-CLA. (B, C) Quantification (percentage of total FA) of the most interesting FAs (TVA, cis9,trans11-CLA, cis6,trans11-18:2, and the suspected trans11,cis13-CLA) after GC-MS analysis of cellular FAMEs extracted from rat hepatocytes in each experimental condition. Data are presented as mean ± SD (n = 4).
These results suggested that siRNA targeting FADS3 significantly suppressed FADS3 mRNA expression, triggered the partial abolition of the native FADS3 protein expression, and specifically reduced Δ11,13-18:2 synthesis from TVA. Conversely, the recombinant FADS3 expression increased the production of the suspected trans11,cis13-18:2.
DISCUSSION
No function has been attributed to the FADS3 gene yet, but it has been postulated that FADS3 may be a new animal FA desaturase. This hypothesis was tested in this study to address the functional role of FADS3. First, transfection of the 1,350 bp open reading frame of the rat FADS3 in COS-7 cells was used to investigate its putative desaturase activity (Fig. 1). Rat FADS1 and FADS2 were concomitantly expressed in this model to validate the positive or negative results obtained with FADS3. Indeed, when incubated with their known FA substrates (20:3 n-6 for FADS1; 18:3 n-3, 18:2 n-6, 18:1 n-9, and 16:1 n-7 for FADS2), FADS1 and FADS2 were specifically able to produce Δ5- and Δ6-desaturated FAs in detectable amounts (Fig. 1 and data not shown). These results are in accordance with previous studies that have used this recombinant model to demonstrate the surprising diversity of Δ6-desaturase substrates (34), including the conversion of 24:5 n-3 into 24:6 n-3, 24:4 n-6 into 24:5 n-6 (24), and 16:0 into 16:1 n-10 (29).
As opposed to these positive results, COS-7 cells expressing FADS3 did not show any desaturase activity toward these common FAs (Fig. 1). These results demonstrated that recombinant FADS3 displayed no Δ5- and Δ6-desaturase activities, consistent with previous reports describing FADS2-null and FADS1-null mice. In these studies, FADS1 or FADS2 deficiency (6, 35, 36) was not rescued by the presence of FADS3 in terms of PUFA composition of tissues. These data and the present study therefore support the idea that FADS3 is not a Δ5- or Δ6-desaturase isoform capable of compensating the absence of FADS1 or FADS2.
To elucidate the function of the FADS3 gene product, many potential FA substrates were unsuccessfully incubated with the COS-7 cell model expressing FADS3. Finally, upon incubation with TVA (Fig. 2), small amounts of a specific octadecadienoic acid were reproducibly synthesized in COS-7 cells expressing FADS3. The use of 13C labeling confirmed its metabolic conversion from the initial [1-13C]TVA substrate. Additional experiments showed that the unknown FA was not synthesized by a FADS3-catalyzed isomerization of RA, which was concomitantly produced by endogenous Δ9-desaturation of TVA (Fig. 3). Evidence is therefore presented that COS-7 cells expressing rat FADS3 specifically gained the ability to introduce a double bond in the carbon chain of TVA to produce the unknown 18:2. Experiments were then carried out to further characterize the chemical structure of this unknown octadecadienoic acid. First, its DMOX derivative mass spectrum (Fig. 4A) demonstrated that it was a Δ11,13-CLA. Then, comparison of its Rt with a commercially available mixture of Δ11,13-CLA standards lacking the trans11,cis13 isomer showed that the cis11,cis13 and trans11,trans13 isomers with Rt significantly higher (+0.6 and +0.9 min, respectively) could be discarded (Fig. 4B). However, uncertainty remained between the cis11,trans13 and trans11,cis13 isomers. Therefore, we analyzed the DMOX derivative mass spectrum of the commercially available cis11,trans13 standard (Fig. 4C) and showed a reproducible difference in the relative abundance of the ions at m/z 250 and 262 amu with the unknown Δ11,13-CLA (Fig. 4A). This result strongly suggested that, based on exclusion of the cis11,trans13 isomer, the unknown Δ11,13-CLA was the trans11,cis13-18:2. In addition, it would be unlikely that the trans-Δ11 double bond preexisting in TVA would be isomerized to a cis double bond at the same time than a trans-Δ13 double bond would be introduced by FADS3. However, because of the lack of commercially available trans11,cis13-CLA standard, the identification could not be considered completely conclusive. Other methodologies, such as GC linked to Fourier transform infrared spectroscopy (FTIR), have been considered for a better characterization of the cis/trans configuration of the product, but FTIR does not distinguish between cis,trans and trans,cis isomers (37) and would not have added information to this characterization. This illustrated the considerable analytical challenge of identifying CLA isomers when well-characterized standards are lacking (30, 37).
Moreover, this unexpected CLA initially characterized in COS-7 cells expressing FADS3 was also found in cultured rat hepatocytes incubated with TVA and 13C-TVA (Fig. 7). Bibliographic data on the trans11,cis13-18:2 (38, 39) revealed that this specific FA has already been identified in the milk and digestas of ruminants (40–46), which are known to produce TVA from dietary linoleic acid (18:2 n-6) by bacteria hydrogenation in their rumen. We also found the presence of this specific CLA in cow milk and goat mammary gland (data not shown). The trans11,cis13-CLA isomer is therefore a naturally relevant FA. In the milk fat from Alpine cows compared with indoor cows, the trans11,cis13-18:2 was greatly increased and represented more than one fourth of total CLA (42), suggesting that its concentration could be related to cows that graze mountain pasture. However, to our knowledge, this particular CLA has not yet been described in humans or in nonruminant species, even after consumption of TVA-enriched diets in studies focusing on the conversion of TVA into rumenic acid (47, 48). In the present study, when rat hepatocytes were exposed to lower concentrations of TVA (50 µM instead of 200 µM, data not shown), the suspected trans11,cis13-18:2 was still detected but represented 0.1% of cell FA (limit of detection). This result suggested that the Δ11,13-CLA may weakly appear under TVA concentrations described as physiological in the plasma of humans (49) and ruminants (50).
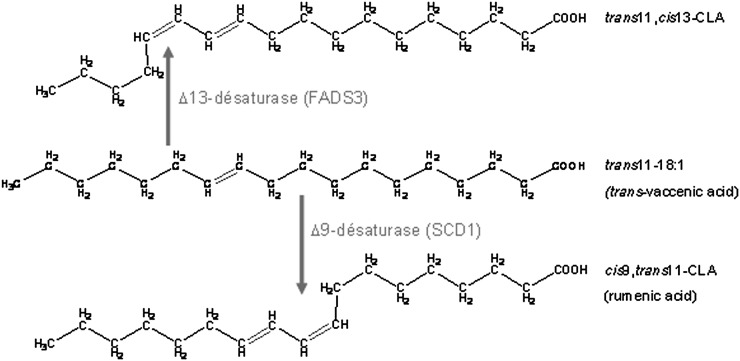
The characterization of this FA as a Δ11,13-CLA means that COS-7 cells expressing rat FADS3 gained the ability to catalyze its synthesis by Δ13-desaturation of TVA. In rat hepatocytes incubated with TVA, we additionally demonstrated that the FADS3 mRNA silencing specifically reduced the synthesis of the suspected trans11,cis13-18:2, whereas conversely, recombinant FADS3 expression increased its production (Figs. 9 and 10). Our report is the first to show that recombinant and native FADS3 are able to introduce a double bond between carbons 13 and 14 of TVA. In previous studies, it has been postulated but never demonstrated that the synthesis of trans11,cis13-18:2 in ruminants could come from dietary 18:3 n-3. Indeed, 18:3 n-3 (cis9,cis12,cis15) can be converted to cis9,trans11,cis15-conjugated triene in the rumen of ruminants (42, 45) and subsequently metabolized to trans11,cis15-18:2 by incomplete bacteria hydrogenation. The last step (i.e., isomerization of trans11,cis15-18:2 to trans11,cis13-18:2) has been suggested but never confirmed. We propose here an alternative new and direct pathway (Fig. 11) catalyzed by FADS3 toward TVA, which is also a product of incomplete bacteria hydrogenation of dietary linoleic acid (18:2 n-6) in the rumen of ruminants. In ruminants, both pathways may coexist, whereas in nonruminant species, including humans, only the FADS3-catalyzed pathway would be possible.
Fig. 11.
Proposed metabolic pathway of trans-vaccenate conversion into cis9,trans11-CLA catalyzed by the Δ9-desaturase and into trans11,cis13-CLA by the newly discovered FADS3-catalyzed Δ13-desaturase.
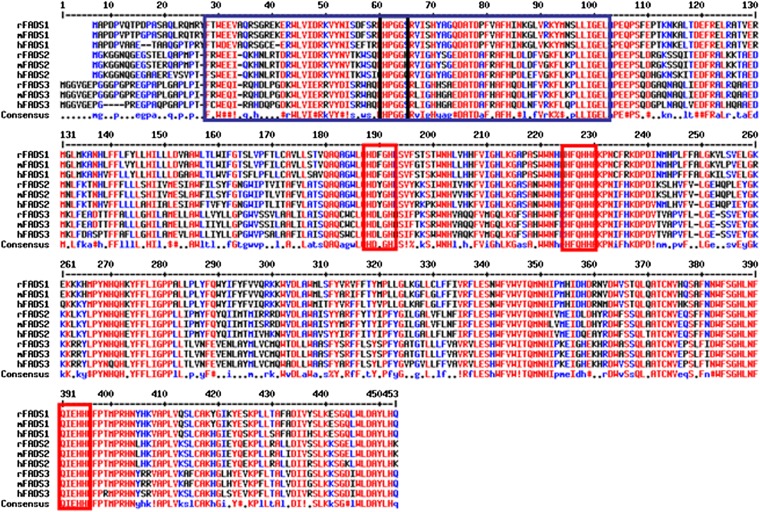
The suspected cis geometrical structure of the newly introduced double bond is not surprising because all of the well-characterized mammalian fatty acyl-CoA desaturases (Δ9-, Δ6-, and Δ5-desaturases) are known to introduce cis double bonds (51). More surprising is the position of the inserted Δ13 double bond, which shows that FADS3 is not a front-end desaturase, as previously hypothesized from its amino acid sequence (14). The so-called group of “front-end” desaturases refers to the desaturases which introduce double bonds between the preexisting double bond (usually located in Δ9 position) and the carboxyl end of a FA (52). The front-end desaturases from eukaryotes are structurally characterized by the presence of an N-terminal cytochrome b5-like domain fused to the main desaturase domain and by the presence of a glutamine (53), which replaces a histidine residue (present in methyl-end desaturases of plants) in the third histidine-rich box H/QX2-3HH, which is the case for FADS3 (11) (Fig. 12). These results show that FADS3 is an exception to the consensus pattern that higher animals, including humans, are devoid of desaturases that insert double bonds in the methyl-end of FA. This exception should be modulated, considering that the first preexisting double bond of the single known FA substrate (i.e., TVA) is a trans-Δ11.
Fig. 12.
Alignment of the amino acid sequences of rat, mouse, and human FADS1, FADS2, and FADS3 deduced from their nucleotide sequences. In the N-terminal end, the cytochrome b5-like domain (present in FADS1, FADS2, and FADS3 sequences) is surrounded by the blue box and characterized by a HPGG motif (black box). In the carboxyl-terminal end, the desaturase domain is characterized by three histidine boxes (red) that are necessary for the catalytic activity, which are present in all the sequences shown.
There has been no report in the literature on a Δ13-desaturase activity in mammals. The existence of such a Δ13-desaturase activity has been described in insects for the multifunctional desaturase from Thaumetopoea pityocampa sex pheromone gland, which converts palmitic acid into (Z)-13-hexadecen-11-ynoic acid (54), but this enzyme presents no sequence homology with FADS3. In a recent study, the metabolic conversion of trans11,trans13-18:2 into cis9,trans11-18:2 in CaCo2 cells suggested the action of an enzyme capable of reducing the Δ13 double bond, after a first Δ9-desaturation step leading to the formation of cis9,trans11,trans13-conjugated linolenic acid (CLnA) as an intermediate (55). We hypothesized that FADS3 could be a Δ13-desaturase/Δ13-reductase bifunctional enzyme, depending on the substrate delivered; however, there was no Δ13-reductase activity when COS-7 cells expressing FADS3 were incubated with α-eleostearic acid (cis9,trans11,trans13-CLnA) (data not shown).
In both COS-7 cells (Fig. 2) and hepatocytes (Fig. 7), the rate of the TVA conversion into the suspected trans11,cis13-18:2 was low, probably because of the enzyme competition of TVA for both Δ9-desaturase and FADS3. This result suggested that the affinity of FADS3 toward TVA was lower than that of SCD1. Although this study focused on FADS3, we also showed that recombinant FADS1 and FADS2 were able to Δ5- and Δ6-desaturate TVA into cis5,trans11-18:2 and cis6,trans11-18:2, respectively (Figs. 2 and 5). Therefore, in the recombinant model, TVA was desaturated by all known animal desaturases (Δ9-, Δ5-, and Δ6-desaturases, and FADS3). In rat hepatocytes, only the products of Δ9-desaturation (cis9,trans11-18:2), Δ6-desaturation (cis6,trans11-18:2), and Δ13-desaturation (trans11,cis13-18:2) were detected (Figs. 7 and 8). Further studies will provide more information on the relative affinity of the animal desaturases for their common TVA substrate in physiological conditions.
To conclude, 13 years after its gene identification in the FADS cluster, this study demonstrates that FADS3 displays a unique and unexpected Δ13-desaturase activity. Inside a large panel of FA, TVA is for now the single substrate to be identified. Therefore, the high selectivity of FADS3 for TVA does not alter the current paradigm of mammalian PUFA dependence on dietary essential FA. Available evidence also indicates that the FADS3-catalyzed Δ13-desaturation of TVA is probably not a major pathway, which is in agreement with its late discovery, but further studies may show that it becomes important in the presence of a high level of dietary TVA (or linoleic acid in ruminants) or under specific physiological conditions. Finally, at present, there is no known function for the suspected trans11,cis13-18:2 product or metabolic basis for thinking that it might be physiologically important. Based on the present description of its FADS3-catalyzed biosynthesis, further studies will probably focus on this particular CLA to answer this question.
Acknowledgments
The authors gratefully acknowledge O. Loreau (CEA Saclay and Cadarache, France) for the supply of [1-13C]-TVA and the Transqual project (ANR-05-PNRA-No.5.E.24). The authors thank P. Toral for the supply of the lipid extracts from goat tissues and F. Boissel for her able technical assistance. The authors also thank S. D'Andréa, who was at the beginning of this long story, and thank J. Wilson-Giron for her English revision of the manuscript.
Footnotes
Abbreviations:
- CLA
- conjugated linoleic acid
- CVA
- cis-vaccenic acid
- DMOX
- 4,4-dimethyloxazoline
- FADS
- fatty acid desaturase
- FAME
- fatty acid methyl ester
- FCS
- fetal calf serum
- NIST
- National Institute of Standards and Technology
- Rt
- retention time
- RA
- rumenic acid
- SCD1
- stearoyl-CoA desaturase 1
- TVA
- trans-vaccenic acid
This work was supported by grants from INRA and Ministère de l'Agriculture.
REFERENCES
- 1.Vanhercke T., Shrestha P., Green A. G., Singh S. P. 2011. Mechanistic and structural insights into the regioselectivity of an acyl-CoA fatty acid desaturase via directed molecular evolution. J. Biol. Chem. 286: 12860–12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi B., Fraser T., Mugford S., Dobson G., Sayanova O., Butler J., Napier J. A., Stobart A. K., Lazarus C. M. 2004. Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nat. Biotechnol. 22: 739–745 [DOI] [PubMed] [Google Scholar]
- 3.Nakamura M. T., Nara T. Y. 2004. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 24: 345–376 [DOI] [PubMed] [Google Scholar]
- 4.Ailhaud G., Massiera F., Weill P., Legrand P., Alessandri J. M., Guesnet P. 2006. Temporal changes in dietary fats: role of n-6 polyunsaturated fatty acids in excessive adipose tissue development and relationship to obesity. Prog. Lipid Res. 45: 203–236 [DOI] [PubMed] [Google Scholar]
- 5.Lauritzen L., Hansen H. S., Jorgensen M. H., Michaelsen K. F. 2001. The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Prog. Lipid Res. 40: 1–94 [DOI] [PubMed] [Google Scholar]
- 6.Stoffel W., Holz B., Jenke B., Binczek E., Gunter R. H., Kiss C., Karakesisoglou I., Thevis M., Weber A. A., Arnhold S., et al. 2008. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 27: 2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquardt A., Stohr H., White K., Weber B. H. 2000. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 66: 175–183 [DOI] [PubMed] [Google Scholar]
- 8.Cho H. P., Nakamura M. T., Clarke S. D. 1999. Cloning, expression, and nutritional regulation of the mammalian Delta-6 desaturase. J. Biol. Chem. 274: 471–477 [DOI] [PubMed] [Google Scholar]
- 9.Cho H. P., Nakamura M., Clarke S. D. 1999. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 274: 37335–37339 [DOI] [PubMed] [Google Scholar]
- 10.Aki T., Shimada Y., Inagaki K., Higashimoto H., Kawamoto S., Shigeta S., Ono K., Suzuki O. 1999. Molecular cloning and functional characterization of rat delta-6 fatty acid desaturase. Biochem. Biophys. Res. Commun. 255: 575–579 [DOI] [PubMed] [Google Scholar]
- 11.Pedrono F., Blanchard H., Kloareg M., D'Andrea S., Daval S., Rioux V., Legrand P. 2010. The fatty acid desaturase 3 gene encodes for different FADS3 protein isoforms in mammalian tissues. J. Lipid Res. 51: 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park W. J., Kothapalli K. S., Reardon H. T., Kim L. Y., Brenna J. T. 2009. Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene. 446: 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillou H., D'Andrea S., Rioux V., Barnouin R., Dalaine S., Pedrono F., Jan S., Legrand P. 2004. Distinct roles of endoplasmic reticulum cytochrome b5 and fused cytochrome b5-like domain for rat Delta6-desaturase activity. J. Lipid Res. 45: 32–40 [DOI] [PubMed] [Google Scholar]
- 14.Rioux V., Pedrono F., Legrand P. 2011. Regulation of mammalian desaturases by myristic acid: N-terminal myristoylation and other modulations. Biochim. Biophys. Acta. 1811: 1–8 [DOI] [PubMed] [Google Scholar]
- 15.Blanchard H., Legrand P., Pedrono F. 2011. Fatty acid desaturase 3 (Fads3) is a singular member of the Fads cluster. Biochimie. 93: 87–90 [DOI] [PubMed] [Google Scholar]
- 16.Brenna J. T., Kothapalli K. S., Park W. J. 2010. Alternative transcripts of fatty acid desaturase (FADS) genes. Prostaglandins Leukot. Essent. Fatty Acids. 82: 281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reardon H. T., Hsieh A. T., Jung Park W., Kothapalli K. S., Anthony J. C., Nathanielsz P. W., Thomas Brenna J. 2013. Dietary long-chain polyunsaturated fatty acids upregulate expression of FADS3 transcripts. Prostaglandins Leukot. Essent. Fatty Acids. 88: 15–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malerba G., Schaeffer L., Xumerle L., Klopp N., Trabetti E., Biscuola M., Cavallari U., Galavotti R., Martinelli N., Guarini P., et al. 2008. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 43: 289–299 [DOI] [PubMed] [Google Scholar]
- 21.Schaeffer L., Gohlke H., Muller M., Heid I. M., Palmer L. J., Kompauer I., Demmelmair H., Illig T., Koletzko B., Heinrich J. 2006. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 15: 1745–1756 [DOI] [PubMed] [Google Scholar]
- 22.Koletzko B., Lattka E., Zeilinger S., Illig T., Steer C. 2011. Genetic variants of the fatty acid desaturase gene cluster predict amounts of red blood cell docosahexaenoic and other polyunsaturated fatty acids in pregnant women: findings from the Avon Longitudinal Study of Parents and Children. Am. J. Clin. Nutr. 93: 211–219 [DOI] [PubMed] [Google Scholar]
- 23.Glaser C., Lattka E., Rzehak P., Steer C., Koletzko B. 2011. Genetic variation in polyunsaturated fatty acid metabolism and its potential relevance for human development and health. Matern. Child Nutr. 7(Suppl. 2): 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Andrea S., Guillou H., Jan S., Catheline D., Thibault J. N., Bouriel M., Rioux V., Legrand P. 2002. The same rat Delta6-desaturase not only acts on 18- but also on 24-carbon fatty acids in very-long-chain polyunsaturated fatty acid biosynthesis. Biochem. J. 364: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zolfaghari R., Cifelli C. J., Banta M. D., Ross A. C. 2001. Fatty acid delta(5)-desaturase mRNA is regulated by dietary vitamin A and exogenous retinoic acid in liver of adult rats. Arch. Biochem. Biophys. 391: 8–15 [DOI] [PubMed] [Google Scholar]
- 26.Rioux V., Lemarchal P., Legrand P. 2000. Myristic acid, unlike palmitic acid, is rapidly metabolized in cultured rat hepatocytes. J. Nutr. Biochem. 11: 198–207 [DOI] [PubMed] [Google Scholar]
- 27.Channing M. A., Simpson N. 1993. Radiosynthesis of 1-[11C] polyhomoallylic fatty acids. J. Labelled Comp. Radiopharm. 33: 541–546 [Google Scholar]
- 28.Loreau O., Maret A., Poullain D., Chardigny J. M., Sebedio J. L., Beaufrere B., Noel J. P. 2000. Large-scale preparation of (9Z,12E)-[1-(13)C]-octadeca-9,12-dienoic acid, (9Z,12Z,15E)-[1-(13)C]-octadeca-9,12,15-trienoic acid and their [1-(13)C] all-cis isomers. Chem. Phys. Lipids. 106: 65–78 [DOI] [PubMed] [Google Scholar]
- 29.Guillou H., Rioux V., Catheline D., Thibault J. N., Bouriel M., Jan S., D'Andrea S., Legrand P. 2003. Conversion of hexadecanoic acid to hexadecenoic acid by rat Delta 6-desaturase. J. Lipid Res. 44: 450–454 [DOI] [PubMed] [Google Scholar]
- 30.Christie W. W., Dobson G., Adlof R. O. 2007. A practical guide to the isolation, analysis and identification of conjugated linoleic acid. Lipids. 42: 1073–1084 [DOI] [PubMed] [Google Scholar]
- 31.Legrand P., Catheline D., Fichot M. C., Lemarchal P. 1997. Inhibiting delta9-desaturase activity impairs triacylglycerol secretion in cultured chicken hepatocytes. J. Nutr. 127: 249–256 [DOI] [PubMed] [Google Scholar]
- 32.Delmonte P., Roach J. A., Mossoba M. M., Losi G., Yurawecz M. P. 2004. Synthesis, isolation, and GC analysis of all the 6,8- to 13,15-cis/trans conjugated linoleic acid isomers. Lipids. 39: 185–191 [DOI] [PubMed] [Google Scholar]
- 33.Chen H., Gu Z., Zhang H., Wang M., Chen W., Lowther W. T., Chen Y. Q. 2013. Expression and purification of integral membrane fatty acid desaturases. PLoS ONE. 8: e58139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guillou H., D'Andrea S., Rioux V., Jan S., Legrand P. 2004. The surprising diversity of Delta6-desaturase substrates. Biochem. Soc. Trans. 32: 86–87 [DOI] [PubMed] [Google Scholar]
- 35.Fan Y. Y., Monk J. M., Hou T. Y., Callway E., Vincent L., Weeks B., Yang P., Chapkin R. S. 2012. Characterization of an arachidonic acid-deficient (Fads1 knockout) mouse model. J. Lipid Res. 53: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stroud C. K., Nara T. Y., Roqueta-Rivera M., Radlowski E. C., Lawrence P., Zhang Y., Cho B. H., Segre M., Hess R. A., Brenna J. T., et al. 2009. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 50: 1870–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De la Fuente M. A., Luna P., Juarez M. 2006. Chromatographic techniques to determine conjugated linoleic acid isomers. Trends Analyt. Chem. 25: 917–926 [Google Scholar]
- 38.Jenkins T. C., Wallace R. J., Moate P. J., Mosley E. E. 2008. Board-invited review: recent advances in biohydrogenation of unsaturated fatty acids within the rumen microbial ecosystem. J. Anim. Sci. 86: 397–412 [DOI] [PubMed] [Google Scholar]
- 39.Shingfield K. J., Bernard L., Leroux C., Chilliard Y. 2010. Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal. 4: 1140–1166 [DOI] [PubMed] [Google Scholar]
- 40.Piperova L. S., Sampugna J., Teter B. B., Kalscheur K. F., Yurawecz M. P., Ku Y., Morehouse K. M., Erdman R. A. 2002. Duodenal and milk trans octadecenoic acid and conjugated linoleic acid (CLA) isomers indicate that postabsorptive synthesis is the predominant source of cis-9-containing CLA in lactating dairy cows. J. Nutr. 132: 1235–1241 [DOI] [PubMed] [Google Scholar]
- 41.Loor J. J., Ueda K., Ferlay A., Chilliard Y., Doreau M. 2004. Biohydrogenation, duodenal flow, and intestinal digestibility of trans fatty acids and conjugated linoleic acids in response to dietary forage:concentrate ratio and linseed oil in dairy cows. J. Dairy Sci. 87: 2472–2485 [DOI] [PubMed] [Google Scholar]
- 42.Kraft J., Collomb M., Mockel P., Sieber R., Jahreis G. 2003. Differences in CLA isomer distribution of cow's milk lipids. Lipids. 38: 657–664 [DOI] [PubMed] [Google Scholar]
- 43.Sehat N., Kramer J. K., Mossoba M. M., Yurawecz M. P., Roach J. A., Eulitz K., Morehouse K. M., Ku Y. 1998. Identification of conjugated linoleic acid isomers in cheese by gas chromatography, silver ion high performance liquid chromatography and mass spectral reconstructed ion profiles. Comparison of chromatographic elution sequences. Lipids. 33: 963–971 [DOI] [PubMed] [Google Scholar]
- 44.Luna P., de la Fuente M. A., Juarez M. 2005. Conjugated linoleic acid in processed cheeses during the manufacturing stages. J. Agric. Food Chem. 53: 2690–2695 [DOI] [PubMed] [Google Scholar]
- 45.Dugan M. E., Kramer J. K., Robertson W. M., Meadus W. J., Aldai N., Rolland D. C. 2007. Comparing subcutaneous adipose tissue in beef and muskox with emphasis on trans 18:1 and conjugated linoleic acids. Lipids. 42: 509–518 [DOI] [PubMed] [Google Scholar]
- 46.Bernard L., Shingfield K. J., Rouel J., Ferlay A., Chilliard Y. 2009. Effect of plant oils in the diet on performance and milk fatty acid composition in goats fed diets based on grass hay or maize silage. Br. J. Nutr. 101: 213–224 [DOI] [PubMed] [Google Scholar]
- 47.Turpeinen A. M., Mutanen M., Aro A., Salminen I., Basu S., Palmquist D. L., Griinari J. M. 2002. Bioconversion of vaccenic acid to conjugated linoleic acid in humans. Am. J. Clin. Nutr. 76: 504–510 [DOI] [PubMed] [Google Scholar]
- 48.Santora J. E., Palmquist D. L., Roehrig K. L. 2000. Trans-vaccenic acid is desaturated to conjugated linoleic acid in mice. J. Nutr. 130: 208–215 [DOI] [PubMed] [Google Scholar]
- 49.Abdelmagid S. A., Clarke S. E., Wong J., Roke K., Nielsen D., Badawi A., El-Sohemy A., Mutch D. M., Ma D. W. 2013. Plasma concentration of cis9trans11 CLA in males and females is influenced by SCD1 genetic variations and hormonal contraceptives: a cross-sectional study. Nutr. Metab. (Lond.) 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen X., Nuernberg K., Nuernberg G., Zhao R., Scollan N., Ender K., Dannenberger D. 2007. Vaccenic acid and cis-9,trans-11 CLA in the rumen and different tissues of pasture- and concentrate-fed beef cattle. Lipids. 42: 1093–1103 [DOI] [PubMed] [Google Scholar]
- 51.Los D. A., Murata N. 1998. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta. 1394: 3–15 [DOI] [PubMed] [Google Scholar]
- 52.Meesapyodsuk D., Qiu X. 2012. The front-end desaturase: structure, function, evolution and biotechnological use. Lipids. 47: 227–237 [DOI] [PubMed] [Google Scholar]
- 53.Sayanova O., Beaudoin F., Libisch B., Castel A., Shewry P. R., Napier J. A. 2001. Mutagenesis and heterologous expression in yeast of a plant Delta6-fatty acid desaturase. J. Exp. Bot. 52: 1581–1585 [DOI] [PubMed] [Google Scholar]
- 54.Abad J. L., Serra M., Camps F., Fabrias G. 2007. Synthesis and use of deuterated palmitic acids to decipher the cryptoregiochemistry of a Delta13 desaturation. J. Org. Chem. 72: 760–764 [DOI] [PubMed] [Google Scholar]
- 55.Schneider A. C., Beguin P., Bourez S., Perfield J. W., 2nd, Mignolet E., Debier C., Schneider Y. J., Larondelle Y. 2012. Conversion of t11t13 CLA into c9t11 CLA in Caco-2 cells and inhibition by sterculic oil. PLoS ONE. 7: e32824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sebedio J. L., Juaneda P., Dobson G., Ramilison I., Martin J. C., Chardigny J. M., Christie W. W. 1997. Metabolites of conjugated isomers of linoleic acid (CLA) in the rat. Biochim. Biophys. Acta. 1345: 5–10 [DOI] [PubMed] [Google Scholar]
- 57.Banni S., Angioni E., Murru E., Carta G., Melis M. P., Bauman D., Dong Y., Ip C. 2001. Vaccenic acid feeding increases tissue levels of conjugated linoleic acid and suppresses development of premalignant lesions in rat mammary gland. Nutr. Cancer. 41: 91–97 [DOI] [PubMed] [Google Scholar]
- 58.Banni S., Carta G., Angioni E., Murru E., Scanu P., Melis M. P., Bauman D. E., Fischer S. M., Ip C. 2001. Distribution of conjugated linoleic acid and metabolites in different lipid fractions in the rat liver. J. Lipid Res. 42: 1056–1061 [PubMed] [Google Scholar]
- 59.Wolff R. L., Christie W. W. 2002. Structures, practical sources (gymnosperm seeds), gas-liquid chromatographic data (equivalent chain lengths), and mass spectrometric characteristics of all-cis 5-olefinic acids. Eur. J. Lipid Sci. Technol. 104: 234–244 [Google Scholar]
- 60.Hamilton J. T., Christie W. W. 2000. Mechanisms for ion formation during the electron impact-mass spectrometry of picolinyl ester and 4,4-dimethyloxazoline derivatives of fatty acids. Chem. Phys. Lipids. 105: 93–104 [DOI] [PubMed] [Google Scholar]