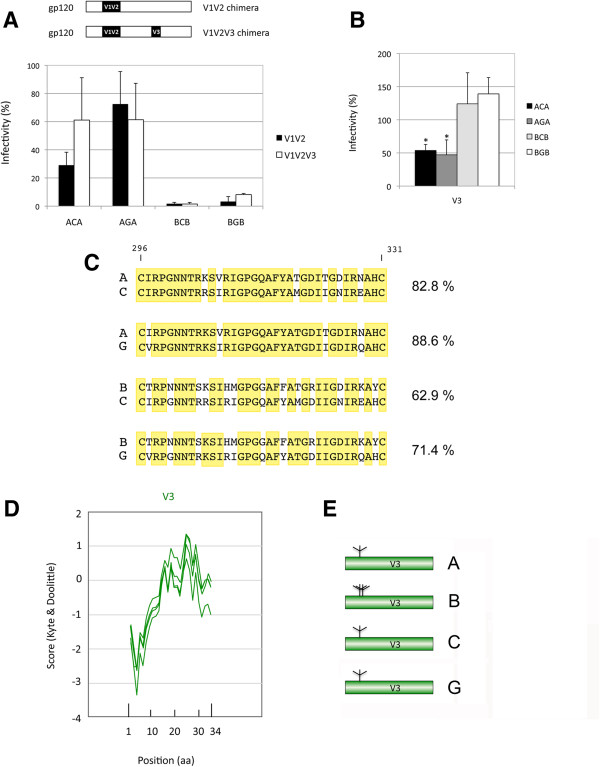
Figure 3.
Effect of the presence of homologous V1V2 and V3 regions on the functionality of the chimeric envelopes.Panel A. Comparison of the functionality of V1V2/V3 chimeras with respect to V1V2 chimeras. The values for the V1V2 chimeras (from Figure 2A) are given as a reference in black. Panel B. Functionality of V3 chimeras. Values are the average of 4 to 5 independent experiments in both panels. The asterisk indicates a significant difference (p < 0.001) in the functionality of the chimera with respect to the corresponding wild-type receiver protein. Panel C. Amino acid sequence alignment of the V3 region for the isolates used to generate the chimeras characterized in Panel B. Identical residues are in a yellow background. The sequences are shown including the C residues that border the V3 region. Alignments were performed using MUSCLE 3.8 [55]. The percentage of identity between the isolates is given on the right of each alignment. Panel D. Hydrophobicity profile of the variable region V3. The profile of the four sequences most intensively studied here (isolates A, B, C, and G) is given. The position in amino acids is given on the x axis, with numbering starting from the first amino acid (N-ter) after the cysteine residue that marks the border of the C2-V3 transition. Panel E. Potential N-Glycosylation sites of the variable region V3 for each subtype used. The location of N-glycosylation sites is indicated. Only sites with a predicted N-glycosylation potential >0.5 according to Server NetNGlyc 1.0 [64] are shown.