Abstract
One of the most popular and simple models for the calculation of pKas from a protein structure is the semi-macroscopic electrostatic model MEAD. This model requires empirical parameters for each residue to calculate pKas. Analysis of current, widely used empirical parameters for cysteine residues showed that they did not reproduce expected cysteine pKas; thus, we set out to identify parameters consistent with the CHARMM27 force field that capture both the behavior of typical cysteines in proteins and the behavior of cysteines which have perturbed pKas. The new parameters were validated in three ways: (1) calculation across a large set of typical cysteines in proteins (where the calculations are expected to reproduce expected ensemble behavior); (2) calculation across a set of perturbed cysteines in proteins (where the calculations are expected to reproduce the shifted ensemble behavior); and (3) comparison to experimentally determined pKa values (where the calculation should reproduce the pKa within experimental error). Both the general behavior of cysteines in proteins and the perturbed pKa in some proteins can be predicted reasonably well using the newly determined empirical parameters within the MEAD model for protein electrostatics. This study provides the first general analysis of the electrostatics of cysteines in proteins, with specific attention paid to capturing both the behavior of typical cysteines in a protein and the behavior of cysteines whose pKa should be shifted, and validation of force field parameters for cysteine residues.
Keywords: MEAD electrostatics model, cysteine pKa calculations, cysteine sulfenic acid
Introduction
In recent years, the importance of cysteine residues to protein function and cellular activity has become increasingly apparent. Cysteines, often due to post-translational modifications including oxidation, nitrosylation, or disulfide bond formation, have been shown to play significant biological roles in enzyme catalysis, in sensing oxidative and nitrosative stress, in the modulation of some transcriptional regulators, in homocysteine binding and in signaling, aging, and disease processes.1–6 The nucleophilic character of cysteine is often an important feature of its reactivity; therefore, stabilization of the deprotonated, thiolate form through lowering of the pKa would promote this reactivity.7 These studies highlight the importance of electrostatics in the mechanism of cysteine reactivity in many cysteine-containing proteins.
As our understanding of the cellular roles that modifiable cysteines can play improves, the ability to accurately predict when cysteines can be modified is becoming increasingly important. Furthermore, there is a need to strike a balance between simple, computationally inexpensive models, which can be used on many protein structures—or even an ensemble of structures—and physically realistic models that can capture wide variation in protein environment.
There have been a few studies of the role of electrostatics in proteins containing active site cysteines7–10; these studies employed simple Poisson–Boltzmann-based models, together with parameters from the literature,11–13 to calculate cysteine pKa shifts and to identify the pKa shift origin. However, in none of these studies was an attempt made either to optimize the cysteine parameters used or to survey the ability of the parameters to capture cysteine behavior across a diverse set of proteins. We do both in this article.
Several different approaches to electrostatic calculations of pKas are available. These range from detailed density functional theory calculations, which can only include portions of the protein, to simpler semi-macroscopic models, which can include the entire protein structure. Most methods involve either single static structures,12,13 or averages over a few structures,11,14 although molecular dynamics with variable protonation states is an emerging, albeit computationally expensive, technique.15–17 The simplest, commonly used semi-macroscopic simple continuum models are applied here.11–13 The general idea of these calculations is to use an implicit water model, such as the Poisson–Boltzmann-based MEAD model, to estimate the pKa shifts of residues in a static structure resulting from electrostatic interactions in the protein environment. The energetics of changing protonation state then involves the electrostatic work needed to alter atom-based charges embedded in an irregularly shaped low dielectric medium (the protein), which is immersed in a high dielectric medium (the solvent).
Four assumptions underpin the semi-macroscopic models: (1) the free energy of ionization can be divided into a local component involving electronic structural changes, for example, bond breaking, and a non-local component which includes distant interactions; (2) model compounds having the same local component as the titratable residues can be used to calculate the local component; (3) the non-local component can be modeled as purely electrostatic; and (4) the electrostatics of the non-local component can be calculated classically using the finite difference linearized Poisson–Boltzmann equation. These assumptions allow for the calculation of an intrinsic pKa, which, when combined with site–site interaction energies, generate full pKas (see Methods for further details). Approximations must be made to calculate the 2N different possible protonation states for N titratable residues, including the reduced-site model,12 Monte Carlo sampling,11 and clustering analyses.18
With adequate parameters and suitable (ad hoc) adjustments of the dielectric constant of the protein, these semi-macroscopic models, along with either reduced-site or Monte Carlo sampling, typically achieve pKa values within ∼ 1–2 pH unit (on average) of experimental results.13,15 Semi-macroscopic methods, especially the MEAD model and package, have been successfully used in a variety of situations.12,13,15,19 Here we employ these methods to: (1) optimize the cysteine parameters to reproduce the expected ensemble behavior across a control group of proteins; (2) determine the ability of the new parameters to reproduce the expected ensemble pKas in a set of proteins whose cysteine pKas are expected to be perturbed; (3) compare calculated and experimental pKa values in a set of cysteine-dependent proteins for which such measurements have been made; and (4) evaluate in detail those proteins for which unexpected pKa values are calculated.
Our long-term goals are to probe the origins of cysteine pKa shifts and to use this information to develop and incorporate electrostatic measures into functional analyses so as to predict reactive cysteine sites in protein sequences. This methodological study is a crucial step toward that goal.
Materials and Methodology
Protein data sets
The control protein set is composed of 60 proteins and 137 cysteine residues (Supporting Information Table 1) and was used for calibrating the cysteine pKa calculations. It was selected to be unbiased and general enough to incorporate a variety of protein folds, with few enough titratable sites to calculate the pKas directly without resorting to Monte Carlo sampling or other more approximate methods. Thus, this control data set was identified as those protein chains in the PDB-Select list20 exhibiting the following features: (1) lacking disulfide bonds; (2) containing fewer than 30 titratable sites; and (3) containing at least one unmodified cysteine residue in the crystal structure. (PDB-Select is used so that each protein exhibits less than 25% pairwise sequence identity to any other; PDB-Select release 2003.20)
The modifiable set of proteins containing cysteine sulfenic acid modification sites was compiled as described elsewhere.21 Cysteines that are modifiable to cysteine sulfenic acid are expected to exhibit a decreased Cys pKa. The modifiable protein set is composed of 47 proteins and 49 modifiable cysteine sites.
Calculation of cysteine pKas
The pKa calculations were performed with the MEAD program suite,11,13 an approach that is semi-macroscopic so that the solvent and the protein have different dielectric constants, taken to be 80 and 20, respectively, in this work, as has been previously determined to be optimal in this model.13 The dielectric boundary is defined as the protein solvent-accessible surface, determined using a water-sized probe radius of 1.4 Å. The linearized Finite Difference Poisson–Boltzmann equations, with an ionic radius of 2.0 Å and ionic strength of 0.15M at 300 K, were used to generate the electrostatic potentials. For each protein in the control and modifiable data sets, a single protein conformation was extracted from the experimental crystal structure and hydrogens were added using the HBUILD facility from CHARMM.22,23 Modified cysteine residues (Cys—SOH or Cys—SO2H) were reduced to the standard sulfhydryl (Cys—SH) form. Structures in which the cysteine had been mutated to a serine for crystallization were computationally changed to cysteine. All structures were subjected to brief restrained minimizations to relieve steric clashes prior to electrostatic calculations; 100 steps of conjugate gradient minimization with force constants of 30 kcal/(mol Å2) on all non-hydrogen atoms, then 100 steps of conjugate gradient minimization with force constants of 20 kcal/(mol Å2) on all non-hydrogen atoms, and finally 100 steps of conjugate gradient minimization with force constants of 10 kcal/(mol Å2) on all non-hydrogen atoms. These minimized proteins were then subjected to the electrostatic calculations.
The reduced site titration model was used to calculate pKas from the electrostatic energies using the MEAD multiflex package11 for the proteins in both the control and modifiable protein sets. For the proteins in the modifiable set with a large (>30) number of titratable sites, where exhaustive calculation was infeasible, a Monte Carlo approach was taken to calculate the pKas.11
Methods for modification of cysteine electrostatic parameters
Every Ser, Asp, Arg, Glu, His, Cys, Lys, Tyr, and Thr residue was considered titratable in the computational model. The model parameters required for these calculations, except for Thr, were originally taken from the parameter files distributed with the MEAD package, along with the radii from the CHARMM22 force field.23,24 These parameters are those developed by Bashford and coworkers in previous studies.25,26 There have been multiple studies using finite-difference Poisson–Boltzmann calculations to study pKa effects with different charge and radii models. These particular parameters were chosen as the most likely consistent parameters as they were developed for use with the CHARMM22 force field models of particular proteins. However, the existing MEAD parameters for the deprotonated Cys produced an unreasonable distribution of pKas for cysteine residues in the control protein set (see Results), hence the need for optimization. Those parameters did come from a different study26 than the majority of the other parameters. The Thr parameters were developed by the current authors in a previous study21 where the particular effects of Thr were found to be important.
The use of CHARMM radii was inspired by previous work25 and by the desire to develop a model for use with MD simulations using the CHARMM force field.16 However, it is possible that the use of different radii may lead to different and perhaps better results or at least to different parameterizations.
It is likely unnecessary to consider all of these residues as titratable as the overall effect of a particular residue on the distribution of another residue's pKas is likely to be modest; however, what we describe here should be a comprehensive set of titratable residues.
Trial charges for the protonated side chain were originally determined using a charge fit to the electrostatic potential obtained from a PM3 calculation,27 a semi-empirical quantum chemical method, on the model compounds using the Gaussian 03 package.28
The charges and model pKas were then modified—both to a single charge model and to distributed charge models based on model PM3 calculations—until a realistic Cys pKa distribution was obtained, that is, one centered around pH 8–8.5. Given the approximate nature of the fitting, it is probable that more accurate parameters could be developed; however, given the paucity of experimental data, this accuracy of the simplest set was considered sufficient. Details of the resulting charge model for cysteine can be found in Table I.
Table I. Cysteine Partial Charge Parameters Recalculated to Give Reasonable Distributions for pKas.
| Residue | Atom | Partial charge (neutral state; atomic units) | Partial charge (charged state atomic units) |
|---|---|---|---|
| Cys | CB | −0.01 | 0.00 |
| Cys | HB1 | 0.09 | 0.00 |
| Cys | HB2 | 0.09 | 0.00 |
| Cys | SG | −0.57 | −1.00 |
| Cys | HG1 | 0.40 | 0.00 |
The table is in standard “.st” format12 for titratable residues. The columns are: atom indicated by CHARMM name, charges of the protonated model, and charges of the unprotonated model.
Results and Discussion
Current parameters do not reproduce expected distributions for Cys pKas
A control set of 60 proteins, containing 137 cysteines in diverse environments, was identified (Supporting Information Table 1). The pKas of the cysteinyl residues in this control set were calculated using standard charge and van der Waals sets associated with MEAD11,12 and the CHARMM22 force field.23,24 We expected these parameters to be able to accurately capture the behavior of an “average” cysteine in a protein.
The initial distribution of cysteine pKas for this set was centered at 10.9, a rather unrealistic mean for a random set of cysteines in proteins (Fig. 1, white bars), where one would expect the mean pKa to range from 8 or 8.5, based on standard textbook values for the free amino acid,29 to 9.5, a pKa based on a “model compound” for Cys30 (where the model compound was OH—CH2—CH2—SH argued to be characteristic of a broad range of alkyl thiols31). Recent experimental measurements of the pKa values of ionizable groups in alanine pentapeptides suggest that 8.55 is a realistic value for Cys pKa.32 This is the first time, to our knowledge, that anyone has demonstrated that a particular parameter set does (or does not) reproduce the average behavior of cysteines in proteins. In our calculations, the pKas of other titratable residues (Ser, Asp, Arg, Glu, His, Lys, and Tyr) exhibit expected distributions, that is, narrow distributions around their standard text book values, without modifications of the parameters, though shifted to the appropriate mean (data not shown).
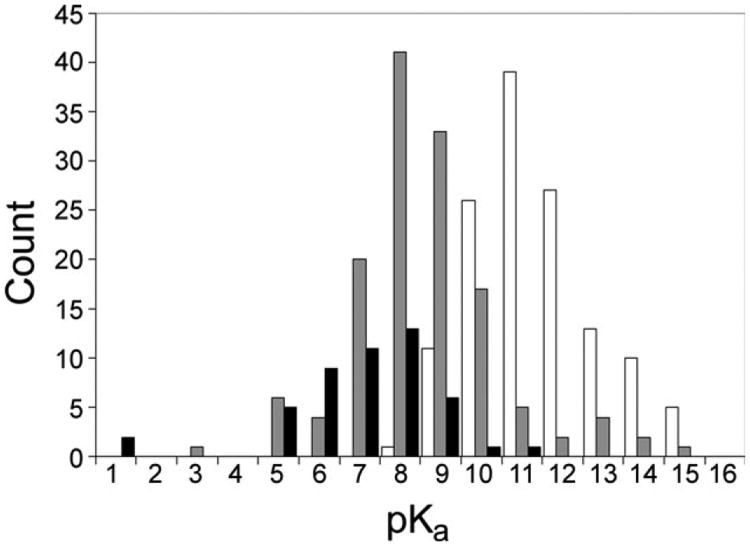
Figure 1.
Distributions of calculated cysteine pKas. The distributions of the calculated cysteine pKas for the control data set with original parameters (white bars), control set with modified parameters (gray bars), and modifiable cysteine pKas (black bars) exhibit means of 10.9, 8.14, and 6.9, respectively. The mean for the control set with modified parameters is consistent with expectations for cysteine pKas, while the mean for the modifiable set is lower, also as expected. The standard deviations are 2.0, 1.9, and 1.4 pH units for the control set with modified parameters, the control set with original parameters, and the modifiable set respectively. Cysteines that lie in the tails of these distributions are discussed in the text.
Modified cysteine parameters reproduce expected Cys pKa distributions
In order to obtain a more realistic distribution, the model compounds' parameters were adjusted (Methods and Table I) until the distribution of the calculated Cys pKas exhibited a reasonable mean of 8.14 (Fig. 1, gray bars), which is in general agreement with textbook values. The distribution exhibits a standard deviation of 2.0 and a standard error of 0.17 and is close to a Gaussian distribution. These results indicate that, overall, calculations with the modified cysteine parameters produce biologically reasonable results. Subsequently, we show that these new cysteine parameters can also capture much of the behavior of reactive cysteines, as well.
In the control set, significant subpopulations exhibit pKas shifted below 7 or above 9 (Fig. 1 and Supporting Information Table 1) and we next explored these outliers. All control set proteins that exhibit a cysteine pKa lower than 5.1 are metalloproteins; furthermore, the observed cysteine is at or near the metal binding site. The proteins with pKas above 12 are all also at or near metal binding sites, except for 2avi, a structure of the avidin-biotin complex. In this complex, the cysteine sulfur atom exhibiting the elevated pKa is 2.04 Å away from the sulfur in a second cysteine, which suggests the presence of an unannotated disulfide bond. If correct, it would make our current calculation of pKa meaningless as the reference state and parameterization would not be appropriate for the disulfide-bonded cysteine. In the remaining structures, the metal ions are not included in the pKa calculations, which suggest that the environment around the metal ions is unusual even in the absence of the metal. This is a promising topic of future study, but is not germane here, except that it suggests that significant shifts in cysteine pKas can be calculated and explained.
Overall, the new cysteine parameters reproduce the expected pKa distribution for a random set of cysteines. Outliers are readily explained as metal binding sites or unannotated disulfide bonds.
Validation of new cysteine parameters by calculation of pKas for modifiable or reactive cysteines
The next validation step is to compare the calculations on another set of proteins where there is biochemical evidence that the cysteine pKa should be decreased compared to those of the control data set. In a separate study,21 a set of proteins was identified that were known to contain a cysteine residue that could be modified to a cysteine sulfenic acid (Cys—SOH). Evidence was based on either biochemical evidence or the presence of this species in the X-ray crystal structure.21 For modification to Cys—SOH by oxidants such as hydrogen peroxide, the reactive form of the cysteine is the deprotonated, negatively charged form. Thus, the pKa of the free, modifiable cysteine residue is expected to be lower than the average.
The cysteine pKa values were calculated for these reactive cysteines in their reduced states. The distribution of calculated pKas for the modifiable cysteines is downward shifted compared to the distribution for pKas of the control set (Fig. 1, compare grayand black bars). The mean pKa of the distribution for modifiable cysteines is 6.9, which is 0.6 standard deviations (using the standard deviation for the control set) lower than the mean for the control set of cysteines of 8.14. (The standard deviations are 1.4 and 2.0 for the modifiable and control sets, respectively.) Overall, this distribution is entirely consistent with the expected enhanced reactivity of these sites.1
While the ensemble distribution for modifiable cysteines is appropriate, we looked at some of the outliers, specifically. Only two modifiable cysteines exhibit pKas shifted more than 1.4 pH units above the control group mean of 8.14 (discussed below). Nearly half, 20 cysteines, exhibit pKas shifted more than 1.4 below the mean of the control set (Fig. 1, black bars). In particular, the modifiable cysteines in two protein structures, protein tyrosine phosphatase 1B (1oet) and RNA triphosphatase domain of mRNA capping enzyme (1i9t), exhibit extraordinarily large pKa shifts greater than 7 pH units, with cysteine pKas of less than 1. Of course, at such a pH these proteins would likely unfold and so, a low pKa would never be observed biochemically. Instead the low pKa indicates the extremely low probability of cysteine protonation of either protein in a physiological environment, which is in agreement with experimental observation for phosphatases.26,33,34
Several of the calculated pKa values are higher than 8, including 2 whose pKas were shifted more than 1.4 pH units above the control group mean of 8.14. We asked whether these values were consistent with what is known about these proteins or whether they were too high, suggesting problems with the parameters or calculations. Cysteine deprotonation is a first step in the reaction, so a lower pKa enhances reactivity of cysteines; however, a decreased pKa is certainly not the only factor which influences reactivity. Unfortunately, we are at an early stage in understanding the features of these reactive cysteines.35,36 In principle, there may be some proteins with unperturbed pKa values (∼8.3–8.5) for which other factors dominate enhanced reactivity. In such cases, the ∼9% of deprotonated form present at a physiological pH of ∼7.4 might be sufficient to support their enhanced reactivity. A second potential explanation for calculated pKa values of highly reactive cysteinyl sites would be the error inherent in such calculations (of around 1 pH unit) that could lead to overestimation of this value.
A third potential explanation is that a different mechanism of Cys—SOH formation, such as oxidation upon exposure to synchrotron radiation, might not require a deprotonated cysteine. Several proteins in the original data set possess Cys—SOH groups that have only been observed in X-ray structures elucidated using synchrotron radiation. In at least some cases, these sites may not be especially reactive toward oxidants such as hydrogen peroxide, but rather, may be sites where hydroxyl radicals generated during exposure of the crystals to intense radiation have caused oxidative damage.37,38 The protein with the highest pKa in this category (at 9.7), with PDB identifier 1vhq (gene name elbB), has not been studied functionally and is tentatively linked to lycopene biosynthesis in E. coli, representing an early step in isoprenoid biosynthesis in these bacteria.39 A more recent computational analysis of the functional sites of this protein suggests that the function is not so clear.40 The other protein, 1hku (C-terminal binding protein 3/BARS, calculated pKa of 9.0), is a corepressor of various transcription regulators involved in the fission of Golgi membranes during mitosis.41,42 This protein is structurally related to a family of 2-hydroxyacid dehydrogenases and possesses NAD+ -dependent dehydrogenase activity. There is no indication in the literature that it undergoes oxidation at cysteine, only that this residue is oxidized in the crystal structure.41 Both proteins have pKas shifted more than 1 standard deviation (1.4 pH units) above the mean of the control set. Other proteins exhibiting unshifted pKas might also contain cysteines modified due to synchrotron radiation damage. 1qvz is another protein of unknown biochemical function in the DJ-1/ThiJ/PfpI superfamily.43,44 This protein is related to the E. coli heat shock protein Hsp31; the putative conserved Cys-His-Glu catalytic triad may therefore be involved in chaperone activity. Two crystal structures have been solved with this Cys (Cys138) oxidized to sulfenic or sulfinic acid, but in both cases this oxidation may have been induced by the synchrotron radiation and the biological relevance of this modification is uncertain.43,44
Qualitatively, the electrostatic calculations on the modifiable cysteine sites are consistent with what is expected: an overall decrease in the pKa of the modifiable cysteines. Further details are consistent with published and experimental information. While the calculations may not calculate the exact cysteine pKa, reactive cysteines for which the pKa is expected to be shifted can be identified.
Comparison to experimentally determined pKas
To further validate the parameters and methods, we studied a set of proteins for which structures are known and pKas were experimentally measured by one or more methods (Table II). The emphasis was on CXXC proteins which, in addition to any biological relevance they may have,9,45 are methodologically interesting as they have two cysteines in close proximity and often occur at the ends of alpha-helices; thus, they present both potentially significant background interactions (from the helix dipole) and residue–residue interactions (from the second cysteine as well as other residues). Cysteine pKas were calculated for this set of proteins and these calculated cysteine pKas were compared with the experimentally determined pKas. As mentioned in the introduction, the results of these calculations are not expected to be more accurate than ∼1–2 pH units, even with a well-developed parameterization.13,15 Only static protein structures are used in these calculations, thus the calculated pKas could potentially be worse if the protein undergoes significant conformational changes around the cysteine sites (discussed subsequently).
Table II. Experimental and Computational Cysteine pKas.
| Proteina | Experimental pKab | PDBc (Res) | Calculated pKa |
|---|---|---|---|
| E. coli Trx46 | 6.7–7.547–50 | 1×0B (Cys32) | 7.6 |
| E. coli DsbA51,52 | 3.4–3.553–54 | 1DSB, 1A2L, 1A2J, 1A2M (Cys30) | 5.5–7.2 |
| E. coli DsbA H32L55 | 4.455 | 1AC1 (Cys30) | 6.6 |
| E. coli DsbA H32S55 | 4.955 | 1ACV (Cys30) | 7.7 |
| E. coli Grx156 | 4.5–<557 | 1EGR (Cys11) | 7.0 |
| E. coli Grx358 | <5.559 | 3GRX (Cys11) | 5.5 |
| Human Grx60 | 3.561 | 1JHB (Cys22) | 5.5 |
| NADH peroxidase62 | <4.563 | 1J0A (Cys42) | 6.7 |
| Human Prx VI64 | 4.6d64,65 | 1PRX (Cys47) | 4.4 |
| Human serum albumin66 | 5.066b,66c | 1N5U (Cys34) | 7.7 |
| Papain67 | 3.3–4.067b,67c | 9PAP (Cys25) | 6.3 |
| Glyceraldehyde-3-phosphate dehydrogenase68 | 5.568b | 1JOX (Cys149) | 4.2 |
Indicates literature reference in which the protein structure is reported.
Indicates literature reference in which the experimentally determined pKa is reported.
PDB file name for the structure on which we calculated the cysteine pKa and the identification of the cysteine for which the pKa was determined.64,65
As referenced by Rhee and coworkers.
For these select proteins, the calculation results are qualitatively correct (Table II). Quantitatively, the mean error is 1.5 pH units, if one averages cases where there are multiple results and if 5.5 and 4.5 are used when the experimental results are <5.5 and <4.5, respectively; thus the new parameters are sufficient to capture the pKa trend. The error is significantly better than from a null hypothesis. The mean error calculated from an “average” cysteine pKa value would be 3.4 pH units using the average cysteine parameterized value of 8.14.
Quantitative agreement between the calculated and experimental pKas is especially significant for the experimental pKas that are particularly low—less than 4.0. The average calculated pKa value across the entire experimental set is 6.3, whereas the average experimental value for this same set is 4.8. This suggests that while our parameters capture the majority of the pKa shifts from average, the methodology does not yet capture the full shift, at least not while accurately modeling a typical cysteine.
One difficulty in this comparison is the lack of agreement in some of the experimental pKa measurements themselves. For instance, we found multiple pKa determinations for the E. coli thioredoxin reactive cysteine in the literature, with measurements ranging from 6.7 to 7.5 (Table II).
Despite the observation that experiment and theory are not in precise quantitative agreement, our current methods are sufficiently accurate to identify the trends toward an increased or decreased pKa and are certainly more accurate than previous parameters. As more pKas are measured and experimental measures of pKas improve, further refinements will be possible. We also expect that if more quantitative accuracy is necessary it might be achieved by combining this model with molecular dynamics simulations.15–17
Electrostatics applied to static crystal structures: the need to include dynamics
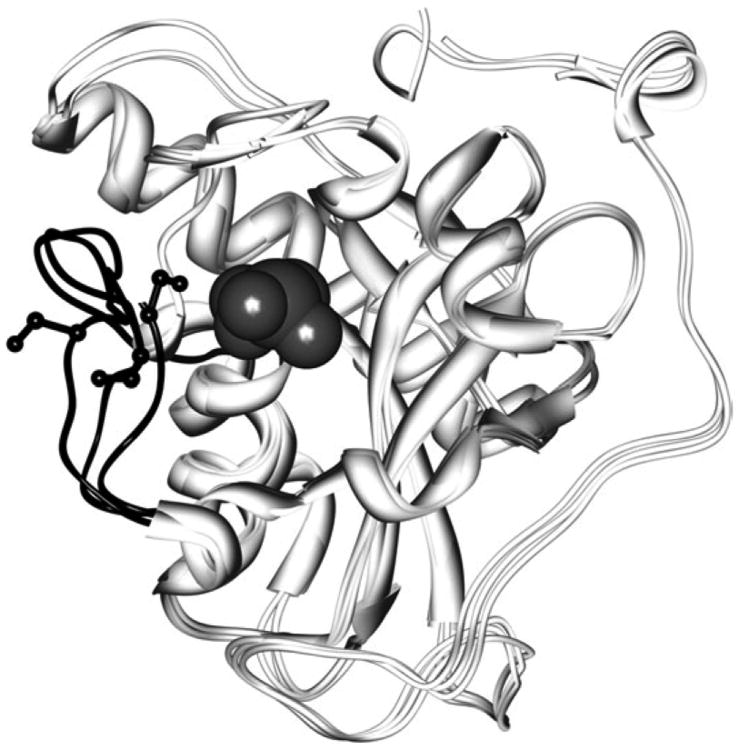
An additional difficulty in computationally reproducing low pKas probably lies with a failure to fully model protein dynamics or conformational changes that may occur as the solution pH is lowered. Several examples are observed here. The protein with the most significantly increased cysteine pKa on the basis of our calculations is methionine sulfoxide reductase (Msr; 1fva69) with a pKa of 10.7. At first glance, this pKa would seem inconsistent with Cys—SOH formation—and this protein does form Cys—SOH. To identify potential reasons for the increased pKa, several Msr structures were compared. Related crystal structures for Msr (1ff369; 1fvg70) show the modifiable cysteine in the same conformation as 1fva; however, a nearby active site loop contains another cysteine in varying conformations (Fig. 2). This loop is exceptionally mobile and is not visible in one crystal structure. pKa values were calculated for the modifiable cysteine in each Msr structure. Depending on the conformation of the loop, the calculated cysteine pKas vary by several pH units, with the pKa of the modifiable cysteine ranging from a low of 8.2 to a high of 10.7 in these static structures. This result illustrates a not unexpected limitation to the current analysis: protein conformational variability, especially with local flexible loops, can modulate the pKa in a manner not captured by a simple model in which conformational relaxation is only modeled via a dielectric constant and with a single structure.
Figure 2.
Comparison of Msr conformations illustrates conformational variability. Three structures of MsrA are superimposed (1fva, 1fvg, and 1ff3), illustrating the conformational variability that exists in the C-terminal region forming an active site loop (shown in black ribbon). This conformational variability affects the pKa of the modifiable cysteines (gray van der Waals spheres). The modifiable cysteines are found in the same conformation in each structure (gray van der Waals spheres), but the nearby active site loop contains another cysteine, found in varying conformations (side chains in black ball and stick representation).
Another set of calculations points to the limitation of applying the simple electrostatics model to a static crystal structure. Calculations on the two modifiable sites in the homodimer from the crystal structure 1prx,64 a peroxiredoxin (Prx), result in different pKas (6.4 and 4.4) due to structural heterogeneity near the modifiable cysteine. While this may provide insight into the subtle local conformational changes that help activate this cysteine, for the purposes of pKa calculation this result again suggests that some conformational averaging, such as through molecular dynamics simulations, 15–17 might be important for producing more accurate calculations in this protein. Of course, it could be that such heterogeneity is real in that the cysteine sites might be nonequivalent enough to give rise to different pKas and presumably different activities (as suggested in recent work16); however, this is far too speculative to conclude from these limited calculations.
Thus, protein dynamics and conformational changes can significantly affect residue pKas. In addition, the conformation does not change as the pH is changed during the calculation. The current calculations are performed on static crystal structures. Although the use of a dielectric constant greater than unity for the protein in the Poisson–Boltzmann calculations assumes that there is a response by the protein to changes in charge states, it does not consider specific changes in conformations that might occur due to specific changes in charges, that is, specific responses by the protein to side chain protonation/deprotonation.
Conclusions
This study provides the first general analysis of the electrostatics of cysteines in proteins, with specific attention paid to capturing both the behavior of a typical cysteine in a protein and the behavior of cysteines whose pKa should be shifted. We have now shown that this behavior can be captured, at least qualitatively, by parameterization of the simple MEAD model through calculations of both “average” and modifiable cysteines. Our results demonstrate that dynamics and conformational changes significantly impact pKa calculations. In at least two proteins observed here, Prx and Msr, there is conformational variability that critically affects the cysteine pKa. Finally, we have shown the limitation of the method in quantitatively predicting pKas that are shifted significantly downward, presumably due to incomplete inclusion of the protein response in the calculation. The parameterization and validation we have conducted will allow for further study of reactive cysteines using the MEAD model with static structures, but also suggests that incorporation of protein conformational changes will be necessary to obtain more quantitative accuracy. Future work will involve probing the origin of cysteine pKa shifts in modifiable cysteines and developing methods to identify and then predict reactive cysteines using these parameters.
Supplementary Material
Acknowledgments
The authors thank Drs. M. Knaggs and W.T. Lowther for helpful discussions, and S. (Knutson) Howerton for assistance with figure preparation. These calculations were performed on Wake Forest University's DEAC cluster, http://www.deac.wfu.edu.
Grant sponsor: (US) NSF; Grant number: MCB-0517343; Grant sponsor: SUR grant from IBM for storage hardware; Grant sponsor: NIH; Grant number: R33 CA126659
Footnotes
Additional Supporting Information may be found in the online version of this article.
The authors have no conflicts of interest to declare.
References
- 1.Wood ZA, Schroder E, Harris JR, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 2.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 3.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 4.Jacob C, Holme AL, Fry FH. The sulfinic acid switch in proteins. Org Biomol Chem. 2004;2:1953–1956. doi: 10.1039/b406180b. [DOI] [PubMed] [Google Scholar]
- 5.Held JM, Gibson BW. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol Cell Proteomics. 2012;11:R111.013037. doi: 10.1074/mcp.R111.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foloppe N, Nilsson L. The glutaredoxin -C-P-Y-C- motif: influence of peripheral residues. Structure. 2004;12:289–300. doi: 10.1016/j.str.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Foloppe N, Sagemark J, Nordstrand K, Berndt KD, Nilsson L. Structure, dynamics and electrostatics of the active site of glutaredoxin 3 from Escherichia coli: comparison with functionally related proteins. J Mol Biol. 2001;310:449–470. doi: 10.1006/jmbi.2001.4767. [DOI] [PubMed] [Google Scholar]
- 9.Moutevelis E, Warwicker J. Prediction of pKa and redox properties in the thioredoxin superfamily. Protein Sci. 2004;13:2744–2752. doi: 10.1110/ps.04804504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warwicker J, Gane PJ. Calculation of Cys 30 delta pKa's and oxidising power for DsbA mutants. FEBS Lett. 1996;385:105–108. doi: 10.1016/0014-5793(96)00358-4. [DOI] [PubMed] [Google Scholar]
- 11.Bashford D. An object-oriented programming suite for electrostatic effects in biological molecules. In: Ishikawa Y, Odehoeft RR, Reynders JVW, Tholburn M, editors. Lecture notes in computer science. Vol. 1343. Berlin: Springer; 1997. pp. 233–240. [Google Scholar]
- 12.Bashford D, Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 13.Antosiewicz J, McCammon JA, Gilson MK. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 14.van Vlijmen HW, Schaefer M, Karplus M. Improving the accuracy of protein pKa calculations: conformational averaging versus the average structure. Proteins. 1998;33:145–158. doi: 10.1002/(sici)1097-0134(19981101)33:2<145::aid-prot1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 15.Lee MS, Salsbury FR, Jr, Brooks CL., III Constant-pH molecular dynamics using continuous titration coordinates. Proteins. 2004;56:738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 16.Yuan Y, Knaggs MH, Poole LB, Fetrow JS, Salsbury FR., Jr Conformational and oligomeric effects on the cysteine pK(a) of tryparedoxin peroxidase. J Biomol Struct Dyn. 2010;28:51–70. doi: 10.1080/07391102.2010.10507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witham S, Talley K, Wang L, Zhang Z, Sarkar S, Gao D, Yang W, Alexov E. Developing hybrid approaches to predict pKa values of ionizable groups. Proteins. 2011;79:3389–3399. doi: 10.1002/prot.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilson MK. Multiple-site titration and molecular modeling: two rapid methods for computing energies and forces for ionizable groups in proteins. Proteins. 1993;15:266–282. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- 19.Koumanov A, Karshikoff A, Friis EP, Borchert TV. Conformational averaging in pK calculations: improvement and limitations in prediction of ionization properties of proteins. J Phys Chem B. 2001;105:9339–9344. [Google Scholar]
- 20.Hobohm U, Scharf M, Schneider R, Sander C. Selection of a representative set of structures from the brookhaven protein data bank. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salsbury FR, Jr, Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 23.MacKerell AD, Jr, Brooks B, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The energy function and its parameterization with an overview of the program. In: Schleyer PvR, Schreiner PR, Allinger NL, Clark T, Gasteiger J, Kollman P., III, editors. The encyclopedia of computational chemistry. Vol. 1. Chichester: Wiley; 1998. pp. 271–277. [Google Scholar]
- 24.MacKerell AD, Jr, Bashford D, Bellott RL, Dunbrack RL, Evanseck JD, Field JJ, Fischer S, Gao J, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DE, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub JE, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 25.You TJ, Bashford D. Conformation and hydrogen ion titration of proteins: a continuum electrostatic model with conformational flexibility. Biophys J. 1995;69:1721–1733. doi: 10.1016/S0006-3495(95)80042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dillet V, Van Etten RL, Bashford D. Stabilization of charges and protonation states in the active site of the protein tyrosine phosphatases: a computational study. J Phys Chem B. 2000;104:11321–11333. [Google Scholar]
- 27.Stewart JJP. Optimization of parameters for semiempirical methods. II. Applications. J Comp Chem. 1989;10:221–264. [Google Scholar]
- 28.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision C02. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 29.Voet D, Voet JG, editors. Biochemistry. 2nd. Wiley, Inc.; New York: 1995. p. 59. [Google Scholar]
- 30.Nozaki Y, Tanford C. Examination of titration behavior. In: Hirs CHW, editor. Methods in enzymology, Vol 11: Enzyme structure. Academic Press; New York: pp. 1967pp. 715–734. [Google Scholar]
- 31.Edsall JT, Wyman J. Biophysical chemistry: thermodynamics, electrostatics, and the biological significance of the properties of matter. Academic Press; New York; 1958. [Google Scholar]
- 32.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein science. 2006;15:1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters GH, Frimurer TM, Olsen OH. Electrostatic evaluation of the signature motif (H/V)CX5R(S/T) in protein-tyrosine phosphatases. Biochemistry. 1998;37:5383–5393. doi: 10.1021/bi971187i. [DOI] [PubMed] [Google Scholar]
- 34.Zhang ZY, Wang Y, Dixon JE. Dissecting the catalytic mechanism of protein-tyrosine phosphatases. Proc Natl Acad Sci USA. 1994;91:1624–1627. doi: 10.1073/pnas.91.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole LB. Formation and function of sulfenic acids in proteins. In: Maines MD, editor. Current protocol in toxicology. New York: Wiley; 2003. pp. 17.11.11–17.11.15. [Google Scholar]
- 36.Poole LB, Hall A, Nelson KJ. Overview of peroxiredoxins in oxidant defense and redox regulation. Curr Protoc Toxicol. 2011;Chapter 7 doi: 10.1002/0471140856.tx0709s49. Unit 7 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu G, Chance MR. Radiolytic modification of sulfur-containing amino acid residues in model peptides: fundamental studies for protein footprinting. Anal Chem. 2005;77:2437–2449. doi: 10.1021/ac0484629. [DOI] [PubMed] [Google Scholar]
- 38.Carugo O, Djinovic Carugo K. When X-rays modify the protein structure: radiation damage at work. Trends Biochem Sci. 2005;30:213–219. doi: 10.1016/j.tibs.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Hemmi H, Ohnuma S, Nagaoka K, Nishino T. Identification of genes affecting lycopene formation in Escherichia coli transformed with carotenoid biosynthetic genes: candidates for early genes in isoprenoid biosynthesis. J Biochem. 1998;123:1088–1096. doi: 10.1093/oxfordjournals.jbchem.a022047. [DOI] [PubMed] [Google Scholar]
- 40.Wei Y, Ringe D, Wilson MA, Ondrechen MJ. Identification of functional subclasses in the DJ-1 superfamily proteins. PLoS Comput Biol. 2007;3:e10. doi: 10.1371/journal.pcbi.0030010. [DOI] [PubMed] [Google Scholar]
- 41.Nardini M, Spano S, Cericola C, Pesce A, Massaro A, Millo E, Luini A, Corda D, Bolognesi M. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 2003;22:3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hidalgo Carcedo C, Bonazzi M, Spano S, Turacchio G, Colanzi A, Luini A, Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- 43.Wilson MA, St Amour CV, Collins JL, Ringe D, Petsko GA. The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: a member of the DJ-1/ThiJ/PfpI superfamily. Proc Natl Acad Sci USA. 2004;101:1531–1536. doi: 10.1073/pnas.0308089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graille M, Quevillon-Cheruel S, Leulliot N, Zhou CZ, Li de la Sierra Gallay I, Jacquamet L, Ferrer JL, Liger D, Poupon A, Janin J, van Tilbeurgh H. Crystal structure of the YDR533c S cerevisiae protein, a class II member of the Hsp31 family. Structure. 2004;12:839–847. doi: 10.1016/j.str.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 45.Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 46.Jeng MF, Campbell AP, Begley T, Holmgren A, Case DA, Wright PE, Dyson HJ. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure. 1994;2:853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- 47.Kallis GB, Holmgren A. Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J Biol Chem. 1980;255:10261–10265. [PubMed] [Google Scholar]
- 48.Chivers PT, Prehoda KE, Volkman BF, Kim BM, Markley JL, Raines RT. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 1997;36:14985–14991. doi: 10.1021/bi970071j. [DOI] [PubMed] [Google Scholar]
- 49.Mössner E, Huber-Wunderlich M, Glockshuber R. Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol-disulfide oxidoreductases. Prot Sci. 1998;7:1233–1244. doi: 10.1002/pro.5560070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mössner E, Iwai H, Glockshuber R. Influence of the pKa value of the buried, active-site cysteine on the redox properties of thioredoxin-like oxidoreductases. FEBS Lett. 2000;477:21–26. doi: 10.1016/s0014-5793(00)01738-5. [DOI] [PubMed] [Google Scholar]
- 51.Martin JL, Bardwell JC, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 52.Guddat LW, Bardwell JC, Martin JL. Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization. Structure. 1998;6:757–767. doi: 10.1016/s0969-2126(98)00077-x. [DOI] [PubMed] [Google Scholar]
- 53.Nelson JW, Creighton TE. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry. 1994;33:5974–5983. doi: 10.1021/bi00185a039. [DOI] [PubMed] [Google Scholar]
- 54.Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 55.Guddat LW, Bardwell JC, Glockshuber R, Huber-Wunderlich M, Zander T, Martin JL. Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci. 1997;6:1893–1900. doi: 10.1002/pro.5560060910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sodano P, Xia TH, Bushweller JH, Bjornberg O, Holmgren A, Billeter M, Wuthrich K. Sequence-specific 1H NMR assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin. J Mol Biol. 1991;221:1311–1324. doi: 10.1016/0022-2836(91)90935-y. [DOI] [PubMed] [Google Scholar]
- 57.Aslund F, Ehn B, Miranda-Vizuete A, Pueyo C, Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nordstrand K, slund F, Holmgren A, Otting G, Berndt KD. NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism. J Mol Biol. 1999;286:541–552. doi: 10.1006/jmbi.1998.2444. [DOI] [PubMed] [Google Scholar]
- 59.Nordstrand K, Aslund F, Meunier S, Holmgren A, Otting G, Berndt KD. Direct NMR observation of the Cys-14 thiol proton of reduced Escherichia coli glutaredoxin-3 supports the presence of an active site thiol-thiolate hydrogen bond. FEBS Lett. 1999;449:196–200. doi: 10.1016/s0014-5793(99)00401-9. [DOI] [PubMed] [Google Scholar]
- 60.Sun C, Berardi MJ, Bushweller JH. The NMR solution structure of human glutaredoxin in the fully reduced form. J Mol Biol. 1998;280:687–701. doi: 10.1006/jmbi.1998.1913. [DOI] [PubMed] [Google Scholar]
- 61.Mieyal JJ, Starke DW, Gravina SA, Hocevar BA. Thioltransferase in human red blood cells: kinetics and equilibrium. Biochemistry. 1991;30:8883–8891. doi: 10.1021/bi00100a023. [DOI] [PubMed] [Google Scholar]
- 62.Yeh JI, Claiborne A, Hol WG. Structure of the native cysteine-sulfenic acid redox center of enterococcal NADH peroxidase refined at 2.8 A resolution. Biochemistry. 1996;35:9951–9957. doi: 10.1021/bi961037s. [DOI] [PubMed] [Google Scholar]
- 63.Claiborne A, Mallett TC, Yeh JI, Luba J, Parsonage D. Structural, redox, and mechanistic parameters for cysteine-sulfenic acid function in catalysis and regulation. Adv Prot Chem. 2001;58:215–276. doi: 10.1016/s0065-3233(01)58006-7. [DOI] [PubMed] [Google Scholar]
- 64.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 65.Kim JR, Yoon HW, Kwon KS, Lee SR, Rhee SG. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 66.a Wardell M, Wang Z, Ho JX, Robert J, Ruker F, Ruble J, Carter DC. The atomic structure of human methemalbumin at 1.9 A. Biochem Biophys Res Commun. 2002;291:813–819. doi: 10.1006/bbrc.2002.6540. [DOI] [PubMed] [Google Scholar]; b Lewis SD, Misra DC, Shafer JA. Determination of interactive thiol ionizations in bovine serum albumin, glutathione, and other thiols by potentiometric difference titration. Biochemistry. 1980;19:6129–6137. doi: 10.1021/bi00567a028. [DOI] [PubMed] [Google Scholar]; c Narazaki R, Hamada M, Harada K, Otagiri M. Covalent binding between bucillamine derivatives and human serum albumin. Pharmaceutical research. 1996;13:1317–1321. doi: 10.1023/a:1016057513490. [DOI] [PubMed] [Google Scholar]
- 67.a Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984;179:233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]; b Lewis SD, Johnson FA, Shafer JA. Potentiometric determination of ionizations at the active site of papain. Biochemistry. 1976;15:5009–5017. doi: 10.1021/bi00668a010. [DOI] [PubMed] [Google Scholar]; c Mellor GW, Patel M, Thomas EW, Brocklehurst K. Clarification of the pH-dependent kinetic behaviour of papain by using reactivity probes and analysis of alkylation and catalysed acylation reactions in terms of multihydronic state models: implications for electrostatics calculations and interpretation of the consequences of site-specific mutations such as Asp-158-Asn and Asp-158-Glu. Biochem J. 1993;294:201–210. doi: 10.1042/bj2940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.a Cowan-Jacob SW, Kaufmann M, Anselmo AN, Stark W, Grutter MG. Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2003;59(Part 12):2218–2227. doi: 10.1107/s0907444903020493. [DOI] [PubMed] [Google Scholar]; b Polgar L. Ion-pair formation as a source of enhanced reactivity of the essential thiol group of D-glyceralde-hyde-3-phosphate dehydrogenase. Eur J Biochem. 1975;51:63–71. doi: 10.1111/j.1432-1033.1975.tb03907.x. [DOI] [PubMed] [Google Scholar]
- 69.Lowther WT, Brot N, Weissbach H, Matthews BW. Structure and mechanism of peptide methionine sulfoxide reductase, an “anti-oxidation” enzyme. Biochemistry. 2000;39:13307–13312. doi: 10.1021/bi0020269. [DOI] [PubMed] [Google Scholar]
- 70.Taylor AB, Benglis DM, Jr, Dhandayuthapani S, Hart PJ. Structure of Mycobacterium tuberculosis methionine sulfoxide reductase A in complex with protein-bound methionine. J Bacteriol. 2003;185:4119–4126. doi: 10.1128/JB.185.14.4119-4126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.