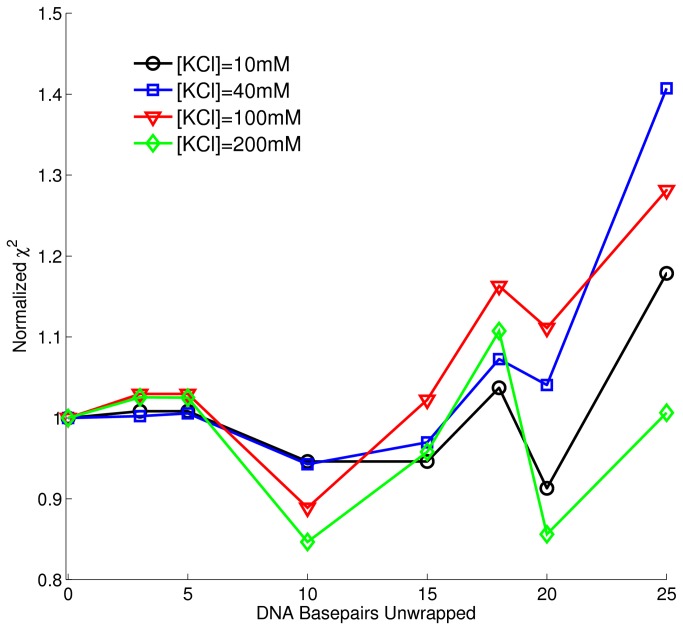
Figure 3. Goodness of fit between gH4 data at multiple salt concentrations and CRYSOL predictions of DNA-unwrapped nucleosomes.
Chi-squared value (goodness of fit) between CRYSOL predicted scattering and SAXS data for the gH4 construct at [KCl]=10mM (circles), 40mM (squares), 100mM (triangles), and 200mM (diamons) with various amounts of DNA basepairs unwrapped. For all salt concentrations, there appears to be two local minima in χ2, one at 10bp unwrapped and one at 20bp unwrapped. The global minimum (best fit) changes from the 20bp unwrapped construct at [KCl]=10mM to the 10bp unwrapped construct at [KCl]=40mM and 100mM. At [KCl]=200mM, there is again a strong shift towards the 10bp unwrapped construct; the 10bp and 20bp χ2 minimum are approximately the same.