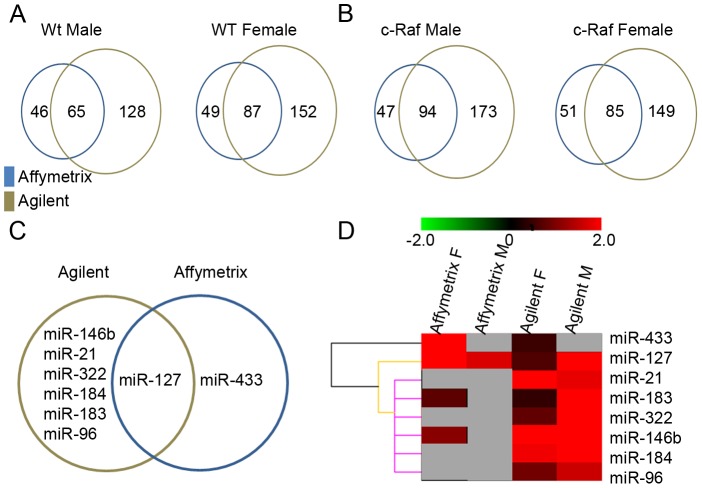
Figure 1. Discordant miRNA expression in lung cancer as determined by the Agilent and Affymetrix microarray platforms.
A) Shown is the Venn diagram of detected miRNAs by the two different microarray platforms in male and female wild-type mice, respectively. B) Shown is the Venn diagram of detected miRNAs by the different microarray platforms in male and female c-Raf transgenic mice, respectively. C) Venn diagram for significantly regulated miRNAs as determined by the different microarray platforms. D) Hierarchical gene cluster analysis. The log2 ratio normalized data were used to perform the HCA analysis for differently expressed miRNAs in the lungs of male and female wild-type and c-Raf-transgenic mice. miR-433 was clearly segregated and found to be exclusively up-regulated in female transgenic mice as detected by the Affymetrix microarray platform. The dendogram is suggestive for miR127 to be regulated in common when both platforms are compared while a third group of miRNAs is regulated in common amongst male transgenic mice as detected by the Agilent platform.