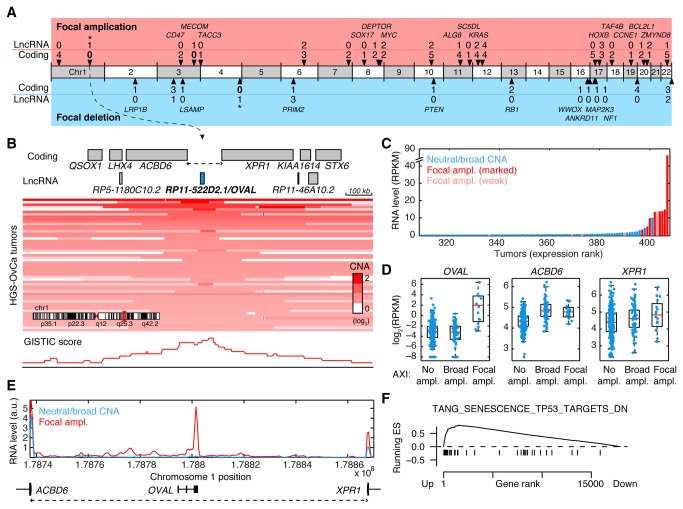
Figure 3. Recurrent lncRNA amplification in serous ovarian carcinoma.
A, LncRNAs and coding genes in narrow regions of recurrent amplification or deletion identified by GISTIC (q < 0.05) in 486 HGS-OvCa tumors. Gene counts for 35 tight focal peaks with a maximum of 5 overlapping coding genes are shown. Known cancer genes and unambiguous coding targets are indicated. *, regions investigated in more detail. B, Detailed view of the ACBD6-XPR1 intergenic region (AXI region, dotted line) in a ~1 Mb genomic context. The AXI focal peak is centered on RP11-522D2.1/OVAL, an uncharacterized lncRNA on chr1q25. Red shading shows copy-number profiles for individual tumors. C, Tumors ordered by OVAL expression. OVAL RNA (y-axis) was mostly low or undetectable, but was induced in focally amplified cases (as defined in Methods). D, Neither ACBD6 nor XPR1 were notably induced by AXI region focal amplification. OVAL RNA was low also in broadly amplified cases. ND, not detected. E, Average RNA-seq read density in the AXI region (dotted line) for tumors with marked AXI focal amplification (n = 10) compared to remaining tumors (normalized read counts per 1000 nt segment). F, GSEA analysis showed than experimentally determined P53 regulated genes are induced in OVAL focally amplified tumors.