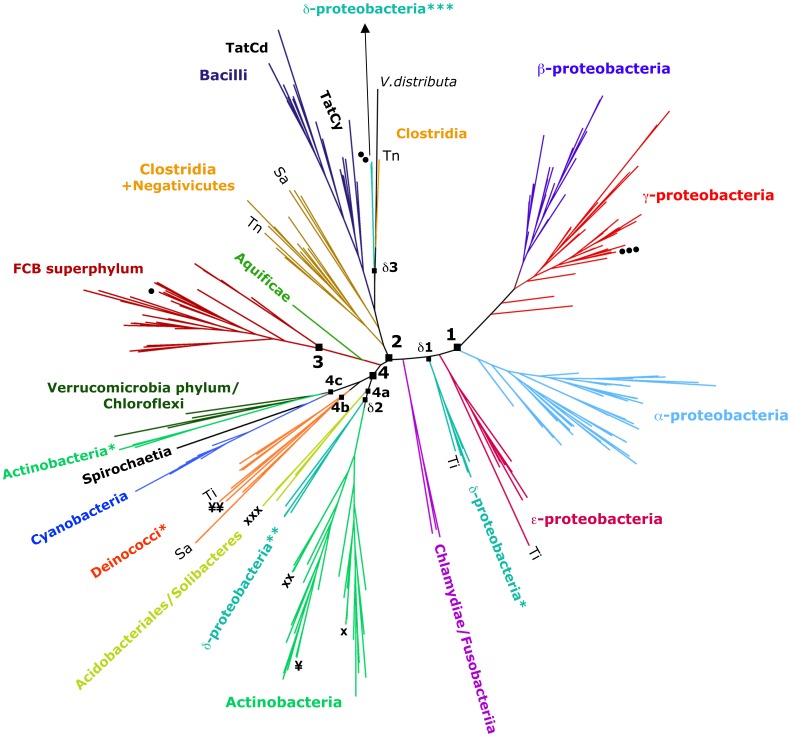
Figure 2. Phylogenetic analysis of TatC protein sequences from diverse bacterial taxa.
The unrooted phylogenetic tree is based on a Bayesian analysis of 232Vulcanisaeta distributa (AC: YP_003902595.1), is labelled as “V.distributa”. Supporting values at each node and individual labels, genus and species names, of all TatC sequences are not shown in this tree for clarity but are provided in the rooted version of the tree in Figure S2. Taxa are described according to their bacterial class, except for classes Verrucomicrobiae and Opitutae (collectively referred to as “Verrucomicrobia phylum”) and classes Chlorobia, Ignavibacteria, Cytophagia, Bacteroidia, Flavobacteria and Sphingobacteria (collectively referred to as “Bacteroidetes/Chlorobi phyla”). Taxa marked with asterisks are detailed as follows: Actinobacteria* includes TatC homologues from class Coriobacteria; Deinococci* includes TatC homologues from classes Deinococci, Thermodesulfobacteria, Nitrospira and Rubrobacteriia; δ-proteobacteria (*) includes TatC homologues from δ-proteobacterial orders Desulfovibrionales and Desulfobacterales; δ-proteobacteria (**) includes TatC homologues from δ-proteobacterial order Myxococcales; δ-proteobacteria (***) includes TatC homologues from δ-proteobacterial order Desulfuromonadales. Symbols adjacent to taxa indicates multiple TatC copies from the same genomes discussed in the Results and Discussion section and the corresponding species are detailed as follows: Cytophaga hutchinsonii (•); Geobacter lovely (••); Colwellia psychrerythraea (•••); Xylanimonas cellulosilytica (x); Streptomyces hygroscopicus (xx); Candidatus Solibacter usitatus (xxx); Nakamurella multipartite (¥); Candidatus Nitrospira defluvii (¥¥); Sulfobacillus acidophilus (Sa); Thermodesulfobium narugense (Tn); Thermodesulfatator indicus (Ti). Labeled nodes are discussed in the Results and Discussion section.