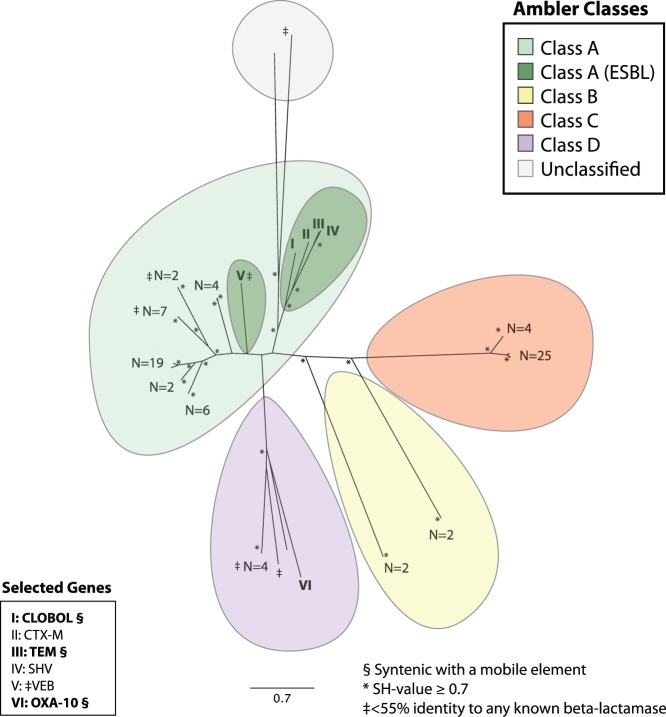
Figure 2. Unrooted approximate maximum-likelihood phylogenetic tree constructed using the predicted amino acid sequences of all beta-lactamases in the study set.
Ambler classes are color-coded as indicated in the legend (upper right). At the terminus of each branch, the number of unique amino acid sequences in the terminal cluster is indicated. Branches with identity to extended-spectrum beta-lactamases (ESBLs) are numbered I–V and their putative ESBL classification is listed in the lower left legend of selected genes. A Class D beta-lactamase with high identity to a known OXA-10 is also numbered (VI) and included in the lower left legend. Genes included in the lower left legend that were syntenic with mobile genetic elements are bolded and marked with a §. Clusters of sequences with <55% ID to any known beta-lactamase are marked with a ‡. Nodes with a Shimodaira-Hasegawa (SH) value > = 0.7 are starred. The Shimodaira-Hasegawa score provides a measure of confidence in tree topology, with a maximum score (highest confidence) of 1.0 [52].