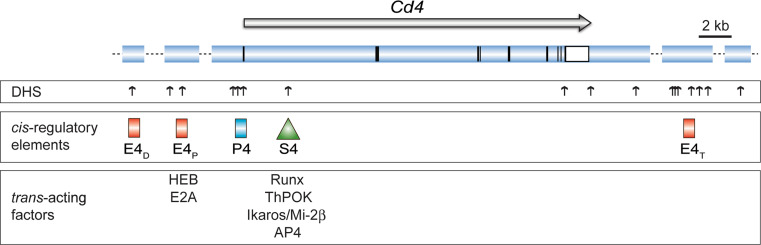
Fig. 2.
Map of the Cd4 gene locus. The upper panel shows the Cd4 gene locus and the horizontal arrow indicates the transcriptional orientation of the Cd4 gene. Vertical black and open bars/squares indicate exons and the 3′ untranslated regions. Vertical arrows indicate DNase I hypersensitivity sites [24–26, 127]. The red rectangles indicate the location of Cd4 enhancers E4D (located approx. 24 kb upstream of the Cd4 promoter), E4P (13 kb upstream), and E4T (20 kb downstream of the 3′ end), while the triangle indicates the location of the Cd4 silencer (S4). Only those trans-acting factors are indicated where binding has been confirmed in primary thymocytes and T cells by chromatin immunoprecipitation assays (see text for more details). Drawing is adapted from reference [71]. The following references report binding to the Cd4 gene locus: Runx complexes [73, 75], ThPOK [64, 128], Ikaros/Mi-2β [89], AP4 [92], HEB/E2A [93]