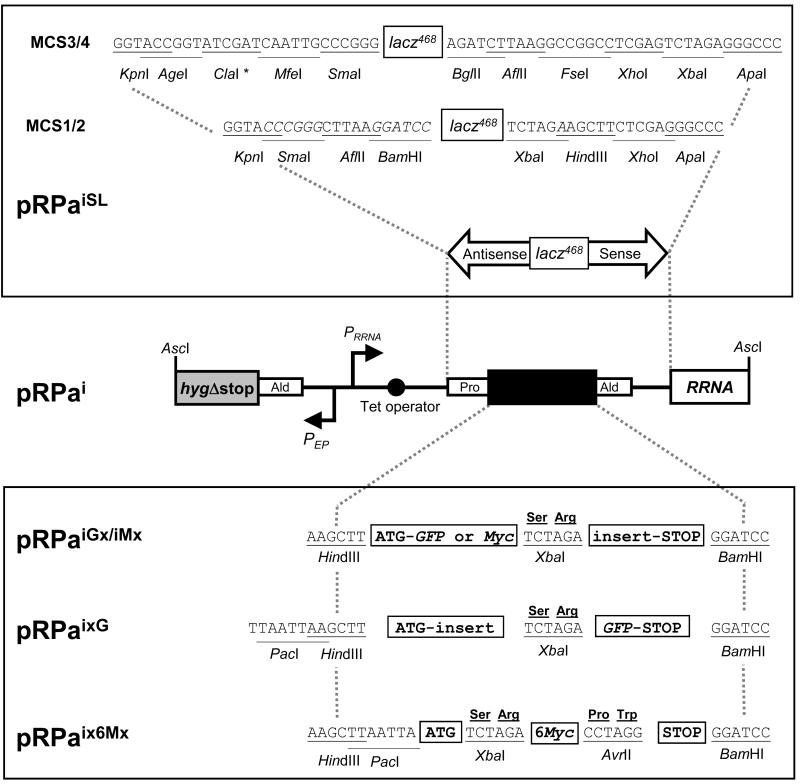
Figure 1. Schematic map of single-locus targeting constructs and available cloning sites.
Following AscI digestion (cleaves GGCGCGCC), the pRPa constructs have terminal hygΔstop and RRNA spacer sequences for targeting the hygΔstart::PAC::RRNA locus, on chromosome 2a in our 2T1 cells. Correct integration generates an intact HYG ORF and deletes a PAC ORF [11]. Selectable marker (HYG) transcription is then constitutive (PEP: procyclin promoter) and independent of the Tet-regulated cassette (PRRNA: RRNA promoter). All cloning sites indicated are unique and several generate cohesive ends that are compatible with other restriction enzymes.
Upper box: Two stem-loop (SL) RNAi constructs are available with different multiple cloning sites (MCS1/2 and MCS3/4). In both cases the ‘loop’ is a 468-bp lacz fragment. We routinely select specific RNAi gene targets (400-600 bp) and primers using the RNAit software [20]
http://trypanofan.path.cam.ac.uk/software/RNAit.html. A single pair of PCR primers are designed that incorporate four selected restriction sites (not present in the RNAi target fragment) such that a single PCR product can be differentially digested and sequentially cloned. For example, using MCS1/2, the following primers could be used to clone antisense followed by sense fragments: Primer 1, XbaI-BamHI-5′ sequence; Primer 2, ApaI-KpnI-3′ sequence. The antisense and sense fragments are generated by BamHI-KpnI and XbaI-ApaI digestion respectively. We’ve seen no problems with SL-construct stability or yield using E. coli DH5α. * The ClaI site in MCS3 is blocked by overlapping Dam methylation in DH5αcells.
Lower box: Tagging constructs are available for expression of N- or C-terminal fusions with eGFP (G, Clontech) or c-Myc (M, EQKLISEEDL). The tandem 6Myc construct allows assembly of either N- or C-terminal fusions and improved sensitivity. x indicates location of selected ORF.
We maintain 2T1 T. brucei cells (Lister 427, MITat1.2, clone 221a), that also express the Tn10 Tet repressor (TetR::BLE::TUB), in HMI-11 growth medium containing phleomycin and puromycin (0.5 μg ml−1 and 0.2 μg ml−1 respectively). We use electroporation (Gene Pulser II, BioRad) with ~2.5 × 107 cells and ~3 μg (yield from one mini-prep, Qiagen) of each AscI-linearised construct in duplicate; each mixture is transferred to 36 ml of medium in a 25 cm2 culture flask. After ~6 h, the appropriate drug selection is applied (2.5 μg ml−1 hygromycin) and cells are distributed into 12- or 24-well plates. Each electroporation experiment typically, and conveniently, yields 2-5 transformed clones using this approach and these are visible after ~5 days (~100,000 cells). To screen clones for simple, double-crossover integration and loss of the PAC gene, we remove ~5% of each day-5, hygR culture and grow with or without puromycin (2 μg ml−1) in a 24-well plate for another 24-48 h. Typically, ~50% of clones are puromycin sensitive and two of these are retained for further analysis. Note that these strains are now resistant to phleomycin (TetR::BLE::TUB) and hygromycin (HYG::RPa::RRNA). Initial analysis normally consists of comparing uninduced and induced (Tet at 1 μg ml−1) cells. For RNAi, this is by counting cell density every 24 h for 3-4 days to generate a growth curve with parallel RNA and/or protein extraction (24 and/or 48 h) for assessment of knockdown by northern and/or western blot depending upon the availability of a suitable antibody. For recombinant protein expression, it is by western blot and/or (immuno)fluorescence analysis after 24 h induction (peak expression is normally seen at this time [10]). Construct maps and sequences, along with further details, are available at http://homepages.lshtm.ac.uk/~ipmbdhor/dhhome.htm (Resources). Further developments and other compatible or complementary constructs will also be documented at this site.