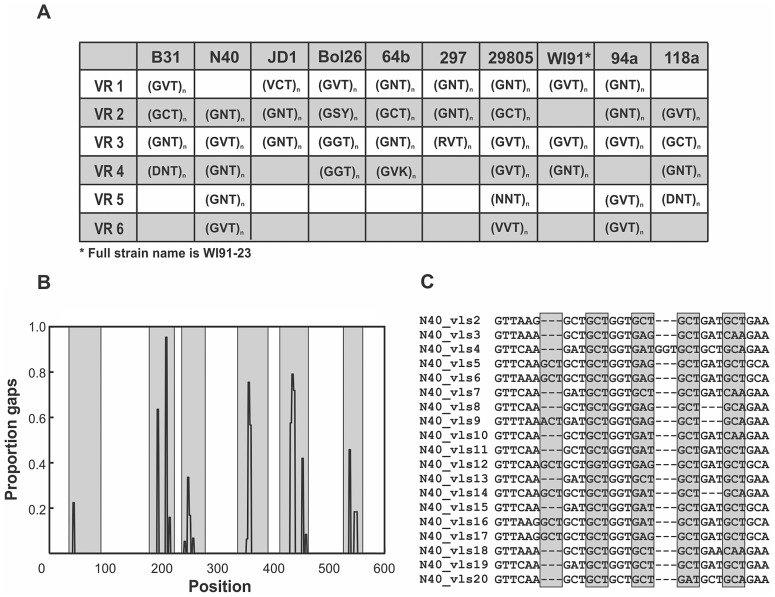
Figure 4. Mutation-prone tandem-repeat motifs are conserved in regions of the unexpressed cassettes that correspond to the antigenic loop domains.
A) Highly-mutable tandem-repeat motifs were identified in the majority of the regions of the cassettes that correspond to the antigenically important loop domains in each strain despite little sequence conservation among strains. Polymorphic sites, denoted with ambiguities following the IUPAC standard, occur primarily in the second codon position. Although the tandem-repeat motifs are similar within the vls unexpressed cassettes of each strain, the number of repeats varies due to insertion and deletion (indel) mutations. B) Tandem-repeat motifs are associated with high frequencies of indel mutations in antigenic loop regions (shaded). The association between tandem repeats and indel mutations in the regions corresponding to the antigenic loop domains is represented as the frequency of sites with gaps in the nucleotide alignment of unexpressed cassettes in strain N40. Indels are present at significantly lower frequencies in antigenic loop regions of the unexpressed cassettes that do not contain tandem repeats (Fig. S1) and are absent in the conserved alpha helical regions that are devoid of tandem-repeat motifs. C) An alignment of antigenic loop region 2 of the N40 unexpressed cassettes illustrates the 3 bp tandem-repeat motifs common to antigenic loop regions of all strains.