Abstract
A synthetic substrate enables a new colorimetric screen for terpene synthase cyclization activity, facilitating the engineering of these enzymes. Using directed evolution, the thermostability of the sesquiterpene synthase BcBOT2 was increased without the loss of other properties. The technique also enabled rapid optimization of conditions for expression and stabilization in lysate of another terpene synthase, SSCG_02150.
Keywords: directed evolution, high-throughput screening, protein engineering, terpenes, thermostability
The development of high-throughput assays can be extremely challenging, yet is essential for many applications in drug discovery and enzyme engineering.[1] Directed evolution has proven to be a reliable method for optimizing the performance of enzymes in a variety of applications,[2] but it requires an appropriate high throughput assay for screening mutant libraries. Terpene synthases catalyze the key cyclization step in the biosynthesis of terpenoids, which are by far the largest class of natural compound and are highly valued as medicines, materials, fuels, and chemicals.[3] Although metabolic engineering efforts have improved access to some terpenoids by production in microbial hosts, the terpene synthase enzymes responsible for the cyclization of linear isoprenoid diphosphates into cyclic terpenoids have proven difficult to engineer.[4] The cyclizations proceed through complex carbocation bond-forming reactions and migrations, and are terminated by either elimination or aqueous quenching of carbocationic intermediates. The complexity of these processes renders rational engineering of terpene synthases nearly impossible. Furthermore, mutagenic studies are reliant upon tedious GC-MS analysis of products, and thus, such studies have explored only small selections of amino acid mutations. The use of directed evolution, therefore, has been difficult because no easy high-throughput assay for productive cyclization activity has been described.
A screen for cyclization activity that provides a colorimetric read-out and can be used in cell lysate would expedite the engineering of terpene synthases by directed evolution and would facilitate the optimization of conditions for soluble, active expression and purification of terpene synthases. We have developed a high-throughput screen that uses a modified substrate containing a vinyl methyl ether functionality and a coupled enzyme assay to detect the cyclization byproduct, methanol. We have used this screen to increase the thermostability of one terpene synthase by directed evolution, and to optimize growth and reaction conditions for a second enzyme.
Several non-natural isoprenoids were shown to serve as substrates for terpene synthases.[5] Notably, Virgil and Corey demonstrated that lanosterol synthase could utilize vinyl ether 1 as a substrate in place of squalene oxide.[5a] In subsequent work by Jin and Coates,[5b] vinyl methyl ether-containing isoprenoid diphosphate 2 was used as a substrate for a diterpene synthase. This vinyl methyl ether, however, hydrolyzed rapidly in neutral aqueous conditions. Inspired by these precedents, we hypothesized that a stable version of a vinyl methyl ether-containing substrate might react in the presence of a terpene synthase to generate methanol as a side product. Thus, we designed substrate 3, which was expected to be less nucleophilic, and therefore less susceptible to hydrolysis, than 2. Substrate 3 was synthesized in four steps from a known compound,[6] and was produced as a mixture of cis and trans isomers. Substrate 3 was stable for at least 10 days at room temperature as a 10 mM stock solution in aqueous 25 mM NH4HCO3.
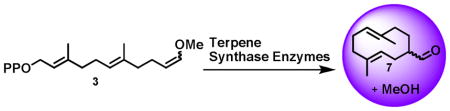
Synthetic substrate 3 has nearly the same length and general shape as farnesyl diphosphate (FPP), the natural substrate for sesquiterpene synthases. Thus, we tested sesquiterpene synthases BcBOT2 and SSCG_02150 for activity on 3 in small-scale reactions using gas chromatography for product analysis.[7] BcBOT2, from the agriculturally important powdery mildew, Botrytis cinerea, produces presilphiperfolan-8β-ol (PSP), a key intermediate in the biosynthesis of the fungus’ virulence factor.[7a] SSCG_02150 is a (−)-δ-cadinene synthase from Streptomyces clavuligerus.[7b] To our satisfaction, these two enzymes generated identical products from 3. According to GC, the product was a mixture of two compounds, which we determined was the result of a thermal Cope rearrangement (caused by the injection port of the GC at 250 °C, see supporting information) of a single product believed to be 7.[8] The terpene synthase-catalyzed reaction that generates 7 is shown in Scheme 1. Upon cyclization of substrate 3, the vinyl methyl ether moiety generates a stabilized carbocation 5, which in turn is quenched by water to generate hemi-acetal 6. Hemi-acetal 6 is unstable and decomposes to the aldehyde 7, releasing one equivalent of methanol in the process.
Scheme 1.
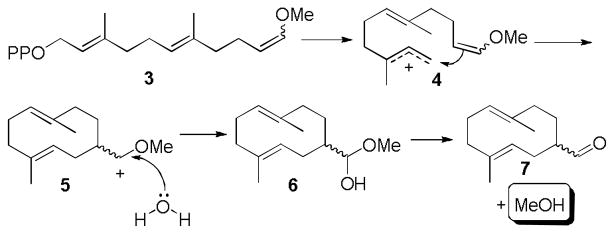
Generation of methanol upon terpen11111e synthase-catalyzed reaction on synthetic substrate 3.
Having shown that these sesquiterpene synthases are able to use 3 as a substrate, we adapted the reaction of 3 for high-throughput screening using an established coupled enzyme assay for methanol.[9d] Substrate 3 and terpene synthase enzymes were allowed to react for as little as 1 min to as much as 24 h in microtiter plates. After the incubation time was reached, alcohol oxidase (AOX), which converts methanol to formaldehyde in the presence of molecular oxygen, was added to the microtiter wells.[9] Following 10 minutes of incubation with AOX, Purpald® was added to react with the formaldehyde to generate a blue-purple color in wells with active enzyme.[10] The absorbance, and corresponding concentration of methanol, was read by a plate reader at 550 nm. Under the conditions used here, concentrations of about 20–250 μM of methanol were found to represent a reasonable working range for the assay, as confirmed by conducting this sequence of AOX and Purpald® treatment with methanol standards (see supporting information). Notably, this screen works well with both purified enzyme and in crude E. coli lysate, and it has the advantage that DTT can be used to stabilize proteins without interfering with the screen response.
Next, the screen was applied to the thermostabilization of terpene synthase BcBOT2 by directed evolution. Thermostable enzymes are much easier to use and are also better starting points for the evolution of new functions. Thermostabilization enables enzymes to operate at higher temperatures, with concomitant faster reaction rates, and allows longer catalyst lifetimes and lower catalyst loadings.[11] Weiss and co-workers undertook the thermostabilization of the plant sesquiterpene synthase, tobacco epi-aristolochene synthase (TEAS), using a computational method, based on the crystal structure of the enzyme, to introduce specific stabilizing interactions.[12] Though the authors were able to improve thermostability, their mutant enzyme was poorly active, insolubly expressed, and the specificity of the product mixture was lost to a large degree (significant side products were formed).
We undertook a directed evolution approach to creating a thermostable terpene synthase with native catalytic activity. A library of about 2800 BcBOT2 mutants was created by error-prone PCR (epPCR) with an average of 3 base pair mutations per gene. This library was heat treated for 10 min at 45°C, and the screen was used to identify mutants that retained activity. Three thermostable mutants were identified. The most improved variant, 19B7, had lost its three C-terminal amino acids and had the mutation K85R. The locations of these mutations within the folded protein structure are not known because no crystal structure of BcBOT2 has been published. The mutations in 19B7 are stabilizing yet do not noticeably change expression, activity, or the specificity for the product (supporting information). The product analysis from GC-scale reactions using the native substrate, FPP, showed only traces of side product formation. Expression was maintained as well. The T50 was increased from 42°C to 47°C. (T50 is the incubation temperature at which 50% of productivity on 3 remained after 10 min of heat treatment.)
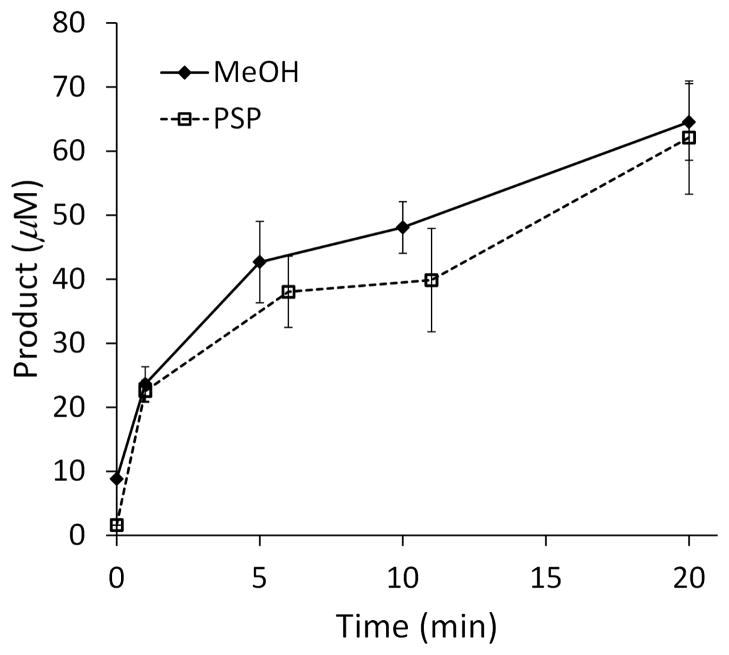
We then confirmed that productivity on the native substrate, FPP, correlated with productivity on 3 as measured by the screen for methanol production. The reaction of purified 19B7 on FPP was followed by GC for 20 minutes, and the amount of cyclization product PSP was quantified at multiple time points. The reaction of 19B7 with 3 was also monitored over 20 minutes under the same conditions. Productivity data with 3 were obtained at time points by terminating the terpene synthase reactions with EDTA, followed by determining the quantities of methanol released using the AOX coupled enzyme assay. As shown in Figure 2, the 19B7-mediated production of methanol from 3 closely follows the cyclization of FPP to make PSP. Importantly, this correlation allows one to use the screen to rapidly estimate the effects of conditions on enzyme productivity instead of using GC-based measurements.
Figure 2.
Correlation of methanol release from 3 (measured by the screen) with presilphiperfolan-8β-ol (PSP) generation from FPP as measured by GC. Reactions were run with 100 nM of thermostable BcBOT2 variant 19B7 at 30°C using 0.20 mM of substrate. Screen background was subtracted from the response.
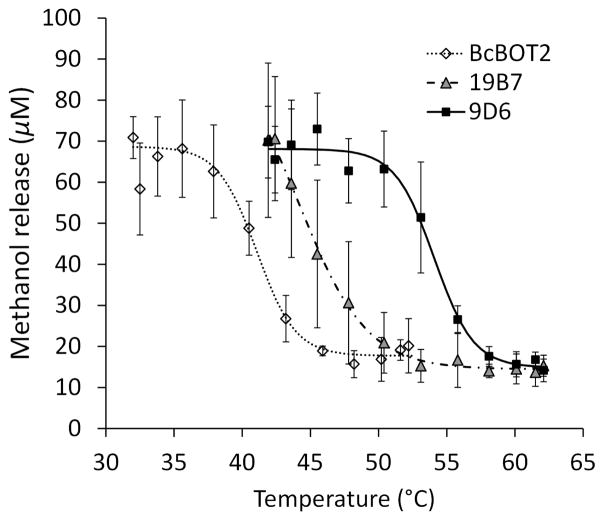
Another round of directed evolution performed on variant 19B7 (again using random mutagenesis with a mutation rate of 3 bp mutations per gene) increased the T50 to about 54°C in variant 9D6 (screening ~1,800 mutants). Variant 9D6 had the mutation H383R, and again product specificity was maintained with the native substrate, FPP, as measured by GC. Expression and activity were also similar to that of BcBOT2. The thermostabilities of 9D6 and 19B7 are shown compared to parent BcBOT2 in Figure 3. In only two rounds of directed evolution and without the advantage of structural knowledge, the thermostability of BcBOT2 was increased by about 12°C with no loss in other desirable properties. Because thermostable proteins are more robust to mutation, 9D6 is an excellent starting point for further functional evolution.
Figure 3.
Methanol release, measured using the screen, of BcBOT2 and mutants after 10 min heat treatment at indicated temperatures, followed by room temperature incubation for 1 h with 3. Error bars indicate standard deviation of at least 3 repeats.
This screening system can also facilitate optimization of expression, purification, and reaction conditions for terpene synthases. Optimization of these properties can be a tedious process because terpene synthases tend to be difficult to express and work with. These enzymes frequently form inclusion bodies or are expressed in inactive form.[13] Even when soluble expression is achieved, purification and reaction buffer conditions must be optimized to keep these sensitive enzymes soluble and active.
We induced protein production in cultures of E. coli containing the SSCG_02150 gene in Luria broth (LB) and terrific broth (TB) at both 18 and 25°C. The resulting cells were lysed with either no buffer additive or with 10% of either sucrose or betaine. The lysates were then treated with 3 for 1 h, and the AOX-Purpald® screen was used to determine which condition(s) gave active enzymes. As shown in Table 1, the screen rapidly revealed that expression in TB was superior to that in LB. Furthermore, expression at 25°C appeared slightly better than at 18°C. Notably, no activity was seen at all for SSCG_02150 in the absence of stabilizing additives, and betaine proved to be the best additive for stabilization. Sucrose appeared to give a slight background response, whereas betaine produced no response above background.
Table 1.
Absorbance values of the screen for various expression conditions.
| LB[a] 18 °C | TB 18 °C | LB 25 °C | TB 25 °C | |
|---|---|---|---|---|
| Blank no additive[b] | 0.20 | 0.17 | 0.17 | 0.17 |
| Blank + betaine | 0.18 | 0.20 | 0.17 | 0.18 |
| Blank + sucrose | 0.22 | 0.24 | 0.22 | 0.24 |
| SSCG_02150 no additive | 0.20 | 0.27 | 0.16 | 0.20 |
| SSCG_02150 + betaine | 0.32 | 0.38 | 0.24 | 0.40 |
| SSCG_02150 + sucrose | 0.26 | 0.38 | 0.22 | 0.22 |
TB is terrific broth with 50 mg/L kanamycin sulfate, LB is Luria broth with 50 mg/L kanamycin sulfate.
Blanks are reported as an average of 2 wells; reactions with SSCG_02150 were reported as an average of 8 wells.
In summary, we have shown that synthetic substrate 3 is effective for screening the cyclization reactions catalyzed by two sesquiterpene synthase enzymes. Using this screen we were able to increase the thermostability of terpene synthase BcBOT2 by 12°C. No structural information was required for this work, and the screen response of thermostabilized mutant 19B7 was predictive for activity on the native substrate FPP. The screen also proved to be a powerful tool for optimizing the expression and buffer conditions for a second sesquiterpene synthase, SSCG_02150. The successful application of this high-throughput screen using synthetic substrate 3 to two distinct enzyme optimization problems suggests that this approach may be generally useful. Optimization challenges for terpenes synthases including overcoming product (inorganic diphosphate) inhibition, increasing activity on native substrates or analogs, altering co-factor (divalent metal cation) preference, and tolerance to organic solvents are all problems that are not obviously solved by rational methods, but may be solved by directed evolution using this screening system.
Experimental Section
Procedure for assay: A microtiter plate with 100 μL per well of cell lysate is treated with 100 μL per well of 0.50 mM substrate 3 in standard buffer (50 mM PIPES, 10 mM MgCl2, 100 mM NaCl, 2 mM DTT, pH 7.5). This solution is made immediately prior to use from a stock solution of 10 mM of substrate 3 in 25 mM aqueous NH4HCO3 (This stock solution is stable for at least 3 months at −20°C). The reactions are incubated at room temp for 1–2 h, and then 10 μL per well of a solution of dilute alcohol oxidase is added (AOX, 50 μL of AOX stock solution from MP Biomedicals dissolved in 950 μL of cold 0.1 M potassium phosphate buffer pH 8.0). The microtiter plate is shaken at room temp for 10 min at 600 rpm on a table top shaker. Optionally, 16 μL of 0.5M EDTA (pH 8.0) can be added to each well to stop the terpene synthase reaction at any point in the process, with shaking for 1 min to mix the reaction contents. Purpald® solution (Aldrich, 351 mg dissolved in 15 mL of 2 N NaOH, 50 μL per well) is added and shaking is resumed for 20–30 min. The plates are briefly centrifuged at 3–5000 rpm to remove bubbles, and the absorbances are read with a plate reader at 550 nm. Background absorbance is typically 0.2 or below, with active enzymes sometimes giving absorbances exceeding 1.0.
Supplementary Material
Figure 1.
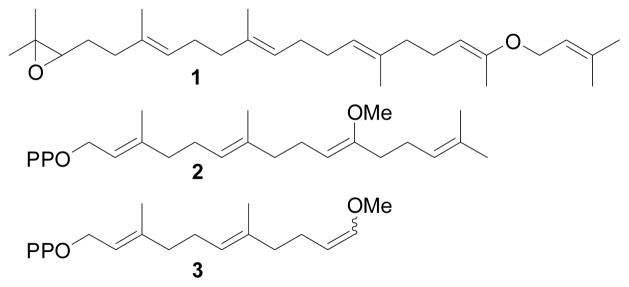
Terpene synthase substrates containing vinyl ether functionalities.
Scheme 2.
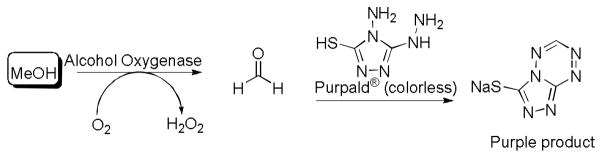
Enzyme-coupled assay for methanol quantification.
Footnotes
We are grateful to David Cane, Scott Virgil, Robert M. Coates, and David Christianson for their pioneering studies, invaluable advice, encouragement, and materials. We thank Indira Wu, Chris Farwell, and Jack Zhang for assistance. RL acknowledges the support of NIH fellowship 1F32GM095061. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. KSR thanks the Deutscher Akademischer Austauschdienst (DAAD) for a postdoctoral fellowship. KZK and RZK acknowledge the support of Summer Undergraduate Research Fellowships from the California Institute of Technology. TH was funded by the FWF grant number: J3327-B21.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.For a compilation of screens, see: Arnold FH, Georgiou G, editors. Directed enzyme evolution: screening and selection methods. Humana Press; Totowa: 2003.
- 2.a) Goldsmith M, Tawfik DS. Curr Opinion Struct Biol. 2012;22:406–412. doi: 10.1016/j.sbi.2012.03.010. [DOI] [PubMed] [Google Scholar]; b) Dalby PA. Curr Opinion Struct Biol. 2011;21:473–480. doi: 10.1016/j.sbi.2011.05.003. [DOI] [PubMed] [Google Scholar]; c) Romero PA, Arnold FH. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Dewick PM. Nat Prod Rep. 2002;19:181–222. doi: 10.1039/b002685i. [DOI] [PubMed] [Google Scholar]; b) Fraga BM. Nat Prod Rep. 2010;27:1681–1708. doi: 10.1039/c0np00007h. [DOI] [PubMed] [Google Scholar]; b) Christianson DW. Curr Opin Chem Biol. 2008;12:141–50. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gao Y, Honzatko RB, Peters RJ. Nat Prod Rep. 2012;29:1153–1175. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Leonard E, Ajikumar PK, Thayer K, Xiao WH, Mo JD, Tidor B, Stephanopoulos G, Prather KLJ. Proc Natl Acad Sci. 2010;107:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Greenhagen BT, O’Maille PE, Noel JP, Chappell J. Proc Natl Acad Sci. 2006;103:9826–9831. doi: 10.1073/pnas.0601605103. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) O’Maille PE, Malone AE, Dellas N, Hess BA, Jr, Smentek L, Sheehan I, Greenhagen BT, Chappell J, Manning G, Noel JP. Nat Chem Biol. 2008;4:617–623. doi: 10.1038/nchembio.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Yoshikuni Y, Ferrin TE, Keasling JD. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]; f) Yoshikuni Y, Martin VJJ, Ferrin TE, Keasling JD. Chemistry and Biology. 2006;13:91–98. doi: 10.1016/j.chembiol.2005.10.016. [DOI] [PubMed] [Google Scholar]; g) Veluda LS, Jiang J, Zakharian T, Cane DE, Christianson DW. Arch Biochem Biophys. 2008;469:184. doi: 10.1016/j.abb.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Corey EJ, Virgil SC. J Am Chem Soc. 1991;113:4025–4026. [Google Scholar]; b) Jin Q, Coates RM. Collect Czech Chem Commun. 2002;67:55–74. [Google Scholar]; c) Hammer SC, Dominicus JM, Syrén PO, Nestl BM, Hauer B. Tetrahedron. 2012;68:7624–7629. [Google Scholar]; d) Siedenburg G, Jendrossekt D, Breuer M, Juhl B, Pleiss J, Sietz M, Klebenberger J, Hauer B. Applied Environ Microbiol. 2012;78:1055–1062. doi: 10.1128/AEM.07059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Chen YL, Chiu HT. J Chinese Chem Soc. 2006;53:1161–1172. [Google Scholar]; f) Faraldos JA, Zhao Y, O’Maille PE, Noel JP, Coates RM. ChemBioChem. 2007;8:1826–1833. doi: 10.1002/cbic.200700398. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Miller DJ, Yu F, Knight DW, Allemann RK. Org Biomol Chem. 2009;7:962–975. doi: 10.1039/b817194g. [DOI] [PubMed] [Google Scholar]; h) Yu F, Miller DJ, Allemann RK. Chem Commun. 2007:4155–4157. doi: 10.1039/b709562g. [DOI] [PubMed] [Google Scholar]; i) Miller DJ, Yu F, Allemann RK. ChemBioChem. 2007;8:1819–1825. doi: 10.1002/cbic.200700219. [DOI] [PubMed] [Google Scholar]
- 6.Labadie GR, Viswanathan R, Poulter CD. J Org Chem. 2007;72:9291–9297. doi: 10.1021/jo7017747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Pinedo C, Wang CM, Pradier JM, Dalmais B, Choquer M, Le Pecheur P, Morgan G, Collado IG, Cane DE, Viaud M. ACS Chem Biol. 2008;3:791–801. doi: 10.1021/cb800225v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yunfeng H, Chou WKW, Hopson R, Cane DE. Chemistry Biology. 2011;18:32–37. doi: 10.1016/j.chembiol.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) de Kraker JW, Franssen CR, de Groot A, Konig WA, Bouwneester H. Plant Physiol. 1998;117:1381–1392. doi: 10.1104/pp.117.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Faraldos JA, Wu S, Chappell J, Coates RM. Tetrahedron. 2007;63:7733–7742. doi: 10.1016/j.tet.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cascon O, Touchet S, Miller DJ, Gonzalez V, Faraldos JA, Allemann RK. Chem Commun. 2012;48:9702–9704. doi: 10.1039/c2cc35542f. [DOI] [PubMed] [Google Scholar]; d) Setzer WN. J Mol Model. 2008;14:335–342. doi: 10.1007/s00894-008-0274-3. [DOI] [PubMed] [Google Scholar]
- 9.a) Sibirny VA, Gonchar MV, Grabek-Lejko D, Pavlishko HM, Csoregi E, Sibirny AA. Intern J Environ Anal Chem. 2008;88:289–301. [Google Scholar]; b) Quesenberry MS, Lee YC. Analytical Biochem. 1996;234:50–55. doi: 10.1006/abio.1996.0048. [DOI] [PubMed] [Google Scholar]; c) Klavons JA, Bennett RD. J Agric Food Chem. 1986;34:597–599. [Google Scholar]; d) Anthon GE, Barrett DM. J Agric Food Chem. 2004;52:3749–3753. doi: 10.1021/jf035284w. [DOI] [PubMed] [Google Scholar]
- 10.Hopps HB. Aldrichimica Acta. 2000;33:28–29. [Google Scholar]
- 11.a) Reetz MT, Carballeira JD, Vogel A. Angew Chem. 2006;118:7909–7915. doi: 10.1002/anie.200602795. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:7745–7751. doi: 10.1002/anie.200602795. [DOI] [PubMed] [Google Scholar]; b) Gumulya Y, Reetz MT. ChemBioChem. 2011;12:2502–2510. doi: 10.1002/cbic.201100412. [DOI] [PubMed] [Google Scholar]
- 12.Diaz JE, Lin C-S, Kunishiro K, Feld BK, Arvantinis SK, Bronson J, Greaves J, Saven JG, Weiss GA. Protein Sci. 2011;20:1597–1606. doi: 10.1002/pro.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Some examples of sesquiterpene synthases: Lin X, Hopson R, Cane DE. J Am Chem Soc. 2006;128:6022–6023. doi: 10.1021/ja061292s.Chou WKW, Fanizza I, Uchiyama T, Komatsu M, Ikeda H, Cane DE. J Am Chem Soc. 2010;132:8850. doi: 10.1021/ja103087w.Garms S, Kollner TG, Boland W. J Org Chem. 2010;75:5590–5600. doi: 10.1021/jo100917c.Cane DE, Watt RM. Proc Natl Acad Sci. 2003;100:1547–1551. doi: 10.1073/pnas.0337625100.Agger S, Lopez-Gallego F, Schmidt-Dannert C. Mol Microbiol. 2009;72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x.Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. J Bacteriol. 2008;190:6084–6096. doi: 10.1128/JB.00759-08.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.