Structures of the ordered domains of the N-proteins.
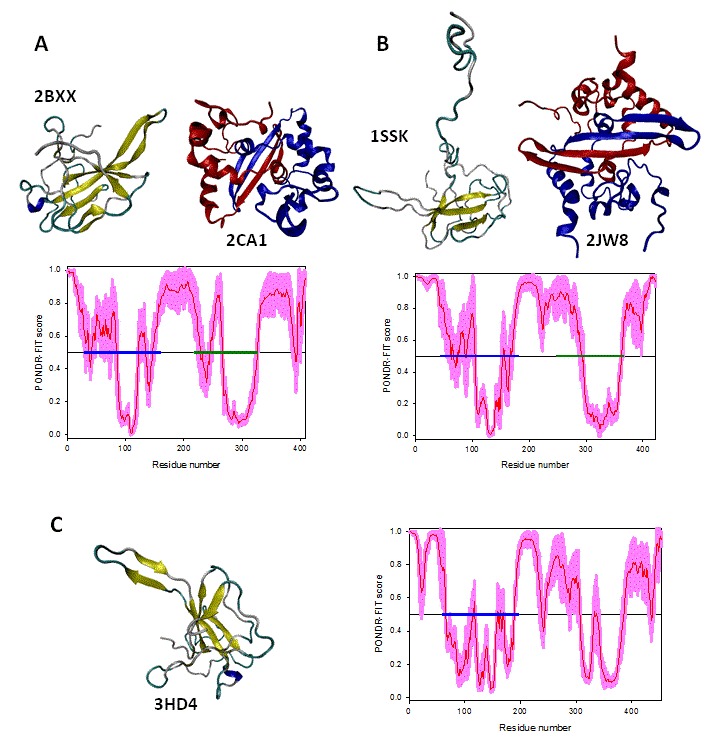
3D-structures of the ordered domains of N-proteins of three coronaviruses from different disorder groups, group A: IBV (A), group B: SARS-CoV (B), and group C: MHV (C). In addition to the available 3D-structures of the RNA-binding and dimerization domains, each panel contains the PONDR-FIT profile, where the localization of these domains is shown by blue and dark yellow bars, respectively. For IBV, the shown structures of the RNA-binding and dimerization domains are 2BXX (residues 29-160) and 2CA1 (residues 218-326). For SARS-CoV, structures of the RNA-binding and dimerization domains are 1SSK (residues 45-181) and 2JW8 (residues 248-365). For MHV, the only available structure is the structure of the RNA-binding domain (PDB code: 3HD4, residues 60-197). Structures were drawn using the VMD software.83
