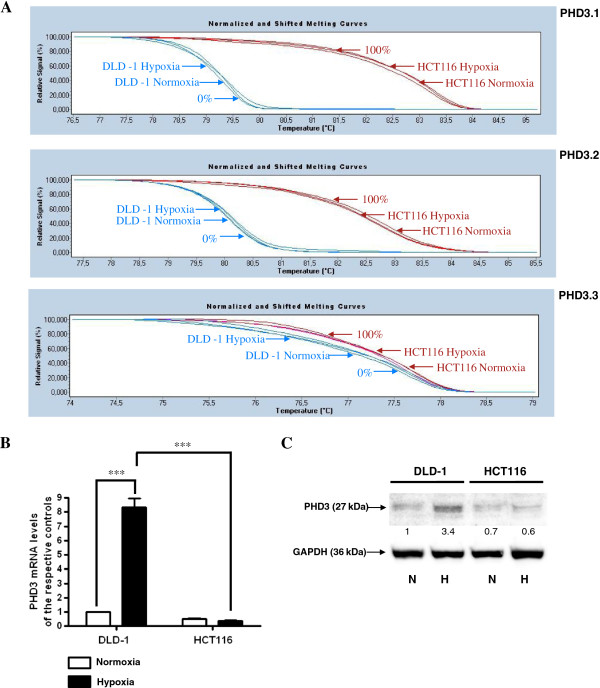
Figure 5.
DNA methylation and expression level of the PHD3 gene in HCT116 and DLD-1 CRC cells. A. HCT116 and DLD-1 cells were cultured under normoxic or hypoxic (1% O2) conditions for 48 hrs. Cells were then used for DNA isolation followed by bisulfite modification. Methylation percentage of three DNA fragments within the PHD3 CpG island (Additional file 1, Additional file 2) in HCT116 and DLD-1 cells under hypoxic and normoxic conditions was determined by Real Time PCR amplification of bisulfite treated standard and cell line DNA, followed by comparison of their HRM profiles. B. Cells were cultured in DMEM either in hypoxic (1%O2) or normoxic conditions for 48 hrs. After incubation, the cells were used for total RNA isolation and reverse transcription. The PHD3 cDNA levels were determined by RQ-PCR relative quantification analysis. RQ-PCR results were standardized by the geometric mean of PBGD and hMRPL19 cDNA levels. PHD3 cDNA levels are expressed as a multiplicity of these cDNA copies in the cell line’s calibrator. C. Cells were cultured in DMEM either in hypoxic (1%O2) (H) or normoxic (N) conditions for 48 hrs. Cells were then used for protein isolation. Proteins were separated by 10% SDS-PAGE, and transferred to a membrane that was then immunoblotted with Rp anti - PHD3 Ab and incubated with goat anti-rabbit HRP-conjugated Ab. The membrane was then stripped and reblotted with Rp anti-GAPDH Ab, followed by incubation with goat anti-rabbit HRP-conjugated Ab. The band densitometry readings were normalized to GAPDH loading control. The ratio of PHD3 to GAPDH for DLD-1 in normoxic conditions was assumed to be 1.