Figure 3.
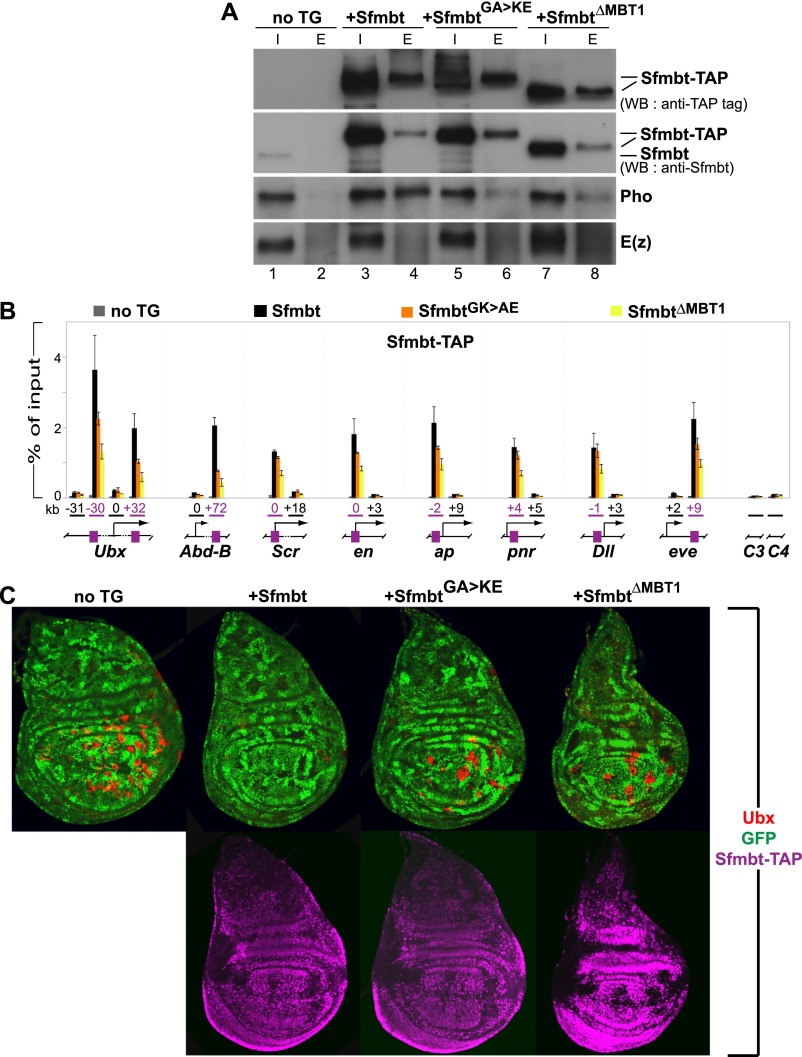
Pho:Sfmbt interaction is critical for Sfmbt function during Drosophila development. (A) Western blot analyses with the indicated antibodies of input (“I”; 2.5% of total) and eluted (“E”; 100% of total) material of IgG sepharose pull-downs of Sfmbt-CTAP proteins from nuclear extracts of 0- to 12-h-old embryos expressing the indicated Sfmbt-CTAP protein or from nontransgenic animals (“not TG”). The tagged proteins were expressed from UASGal4:Sfmbt-CTAP transgenes under the control of the daughterless:Gal4 driver. Blots were first probed with anti-Sfmbt, and then stripped and reprobed with anti-peroxidase antibody to specifically detect the Sfmbt-CTAP fusion proteins. Wild-type Sfmbt-CTAP protein coimmunoprecipitates Pho but not the PRC2-subunit E(z). Significantly lower levels of Pho are coimmunoprecipitated with the SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP proteins (cf. lanes 6,8 and 4). (B) ChIP analysis monitoring binding of the indicated Sfmbt-CTAP proteins at PcG target genes in chromatin of wild-type larvae that express Sfmbt-TAP proteins under the control of the ubiquitous daughterless:Gal4 driver. Graphs show results from three independent immunoprecipitation reactions with anti-peroxidase antibody that binds to the protein A moiety of the TAP tag. ChIP signals, quantified by quantitative PCR, are presented as the mean percentage of input chromatin precipitated at each region; error bars indicate ±SD. Locations of PREs (purple boxes) and other regions relative to the transcription start sites are indicated in kilobases; control regions C3 in euchromatin and C4 in heterochromatin are located remotely from PcG target genes. Sfmbt-CTAP is specifically enriched at PREs. Levels of SfmbtG635K/A638E-CTAP protein binding are reduced at several PREs, whereas binding of SfmbtΔMBT1-CTAP is reduced twofold to fivefold at all analyzed PREs. (C) Wing imaginal discs from third instar larvae stained with antibody against the HOX protein Ubx (red, top row) and anti-peroxidase antibody to detect the Sfmbt-CTAP proteins (purple, bottom row). Clones of Sfmbt1 homozygous cells are marked by the absence of GFP (green) and were induced in animals lacking a transgene (“no TG”) or expressing the indicated Sfmbt-CTAP proteins under the control of the 69B:Gal4 driver. For unknown reasons, the SfmbtΔMBT1-CTAP is expressed at higher levels (see Supplemental Fig. S8) than the Sfmbt-CTAP and SfmbtG635K/A638E-CTAP proteins. Note that only Sfmbt1 homozygous cell clones (“no TG”) in the wing pouch but not in the notum or hinge show strong misexpression of Ubx, as described previously (Klymenko et al. 2006). To evaluate the capacity of the transgene-encoded Sfmbt proteins to repress Ubx in Sfmbt1 mutant cell clones, we therefore only analyzed mutant clones in the wing pouch area for the presence of Ubx protein. For each genotype, multiple wing imaginal discs were analyzed, and in animals expressing a CTAP fusion protein, only clones in which the fusion protein was detected by immunofluorescence labeling (shown in the bottom panel) were scored. In the “no TG” animals, 94% of Sfmbt1 homozygous clones (n = 98 clones) show misexpression of Ubx. In Sfmbt-CTAP animals, repression of Ubx is rescued in most Sfmbt1 homozygous clones, and only 4% of the clones (n = 78 clones) show misexpression of Ubx. In SfmbtG635K/A638E-CTAP animals, 81% of Sfmbt1 homozygous clones (n = 97 clones) show misexpression of Ubx. In SfmbtΔMBT1-CTAP animals, 87% of the clones (n = 31 clones) show misexpression of Ubx. The large proportion of Ubx-expressing Sfmbt1 mutant clones in SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP animals suggests that these two proteins are largely nonfunctional in Polycomb repression.