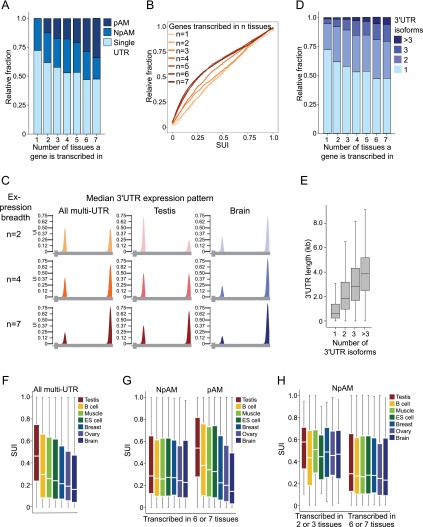
Figure 4.
Ubiquitously transcribed genes are enriched in multi-UTR genes and show significantly less usage of their proximal pA sites than tissue-restricted genes. (A) Fraction of single, NpAM, or pAM genes stratified by the number of tissues in which a gene is transcribed. (B) Cumulative distribution of the SUI of all multi-UTR genes stratified by the number of tissues in which a gene is transcribed. The distribution of the SUI between ubiquitous and tissue-restricted genes is significantly different (Kolmogorov-Smirnoff [KS] test, P < 10−16). (C) Illustration of the median 3′ UTR isoform expression pattern of multi-UTR genes that generate two isoforms stratified by the number of tissues in which a gene is transcribed (expression breadth). (Left panel) Median UI for all multi-UTR genes transcribed in two, four, or seven tissue samples. (Middle panel) Median UI for the same genes in the testis. (Right panel) Median UI for the same genes in the brain. Only values for the most proximal and distal pA sites are shown. A gene model depicting the terminal exon is shown below. (D) Fraction of genes with one, two, three, or more than three 3′ UTR isoforms stratified by the number of tissues in which a gene is transcribed. (E) Distribution of 3′ UTR length stratified by the number of 3′ UTR isoforms observed for a gene. Box plots were plotted as in Figure 3D. (F) Distribution of the SUI of all multi-UTR genes expressed in a given tissue. Box plots were plotted as in Figure 3D. (G) Distribution of the SUI of ubiquitously expressed NpAM and pAM genes. pAM genes were identified using an FDR-adjusted P < 0.1, whereas the shown NpAM genes have an FDR-adjusted P > 0.6. (H) Distribution of the SUI of tissue-restricted and ubiquitously expressed NpAM genes. NpAM genes were identified using an FDR-adjusted P > 0.6. The significance of the difference in the SUI distributions has P-values that range between P = 0.002 and P = 10−9 (Mann-Whitney test), depending on the tissue. Box plots were plotted as in Figure 3D.