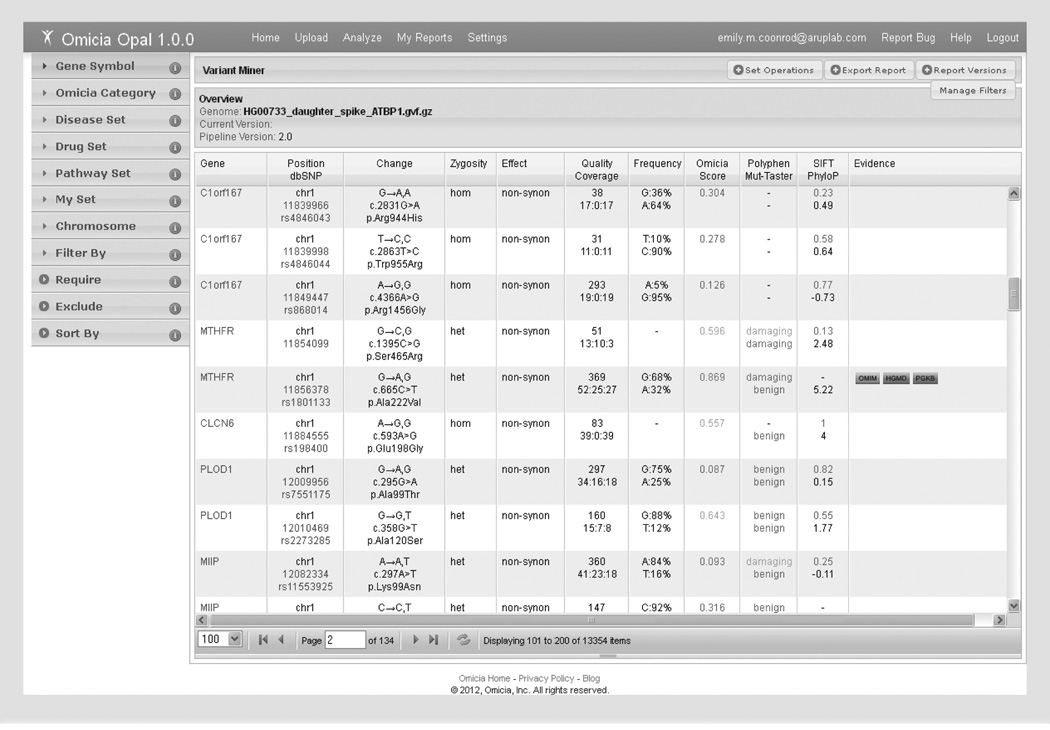
Figure 2. Opal Variant Miner webpage.
The Variant Miner consists of the variant annotation grid and filtering options by knowledge sets or variant properties. To the left of the table are the multiple available filtering options (in the collapsible windows). The bottom of the table lists the number of variants (items) left after each filtering step. Each variant is listed per table row, and ordered numerically by chromosome number and position. Hyperlinks to additional information are available for the Gene (in blue), Position/dbSNP (in blue), and Evidence (boxed) columns. Quality and Coverage information comes from the next-generation sequencing data file, if available. The allele frequency is from the 1000 Genomes frequency data. Red numbers and words in the Omicia/Polyphen/SIFT columns indicate predicted damaging variants, yellow indicates the prediction of a potentially damaging variant and green indicates a benign variant.