Fig 1.
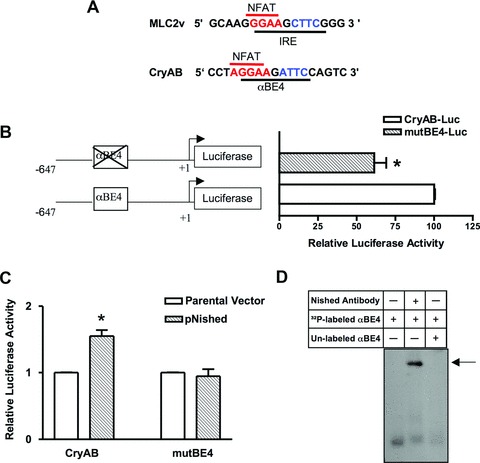
αBE4 enhancer activity. (A) Comparison of sequences of cardiac specific elements, IRE in the MLC2v and the αBE4 in the CryAB gene. The IRE and the αBE4 sequences contain a NFAT binding site, shown in red, 5′ to the Nished binding element, shown in blue. (B) Schematic representation of WT (CryAB-Luc) and mutant (mutBE4-Luc) gene constructs. Numbers denote the position of 5′ and 3′ ends of the gene relative to the transcription initiation site. HL-1 cardiomyocytes were transfected with CryAB-Luc or mutBE4-Luc plasmids. (C) HL-1 cells were co-transfected with CryAB-Luc or mutBE4-Luc plasmids along with parental vector (pcDNAV5) or Nished expression plasmid (pcDNAV5-Nished). Luciferase activities are expressed relative to the mean value derived from cells transfected with CryAB-Luc plasmids (B). In cells co-transfected with pcDNAV5-Nished, luciferase activities are expressed relative to the mean value derived from cells cotransfected with parental vector (pcDNAV5) and CryAB-Luc or mutBE4-luc plasmids. Renilla luciferase activity was used for normalization of transfection efficiency. Experiments have been repeated at least three times. Data are mean ± SE. *P < 0.05 and **P < 0.01. (D) Nuclear extracts from heart tissue of WT mice were incubated with a 32P-labeled oligonucleotide encompassing the αBE4 sequence and analyzed by gel mobility shift assay. Fifty-fold of unlabeled αBE4 DNA was used as a specific competitor. Preincubation of cardiac nuclear extracts with anti-Nished antibody shows a supershift indicated by arrow (n= 3).
