FIGURE 2.
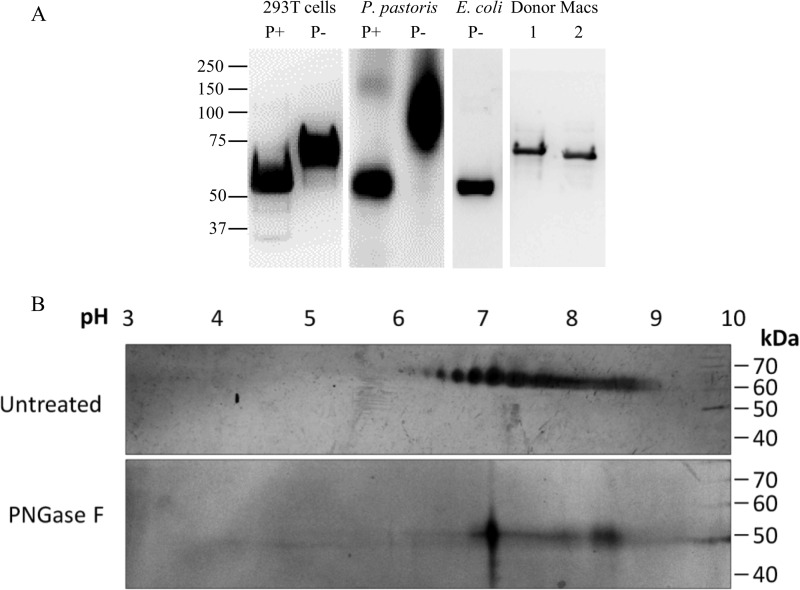
N-Glycosylation altered the molecular mass and biochemical properties of LILRA3. A, Western blotting of PNGase F-treated (P+) and PNGase F-untreated (P−) rLILRA3 from 293T cells and P. pastoris using anti-LILRA3 mAb showed a substantial reduction in molecular mass of both eukaryotic cell-produced recombinant proteins following deglycosylation. Recombinant LILRA3 produced in E. coli served as a control for non-glycosylated protein. Non-PNGase F-treated LILRA3 from native macrophages (Macs) of two individual donors (lanes 1 and 2) is shown as positive references to optimally glycosylated protein. B, silver staining of two-dimensional gel of non-deglycosylated purified rLILRA3 from 293T cells showed a spectrum of isoelectric focusing with pI ranging from 6 to 9 (upper panel), but upon deglycosylation using PNGase F, it was reduced to a single focus with a pI of 7 (lower panel).