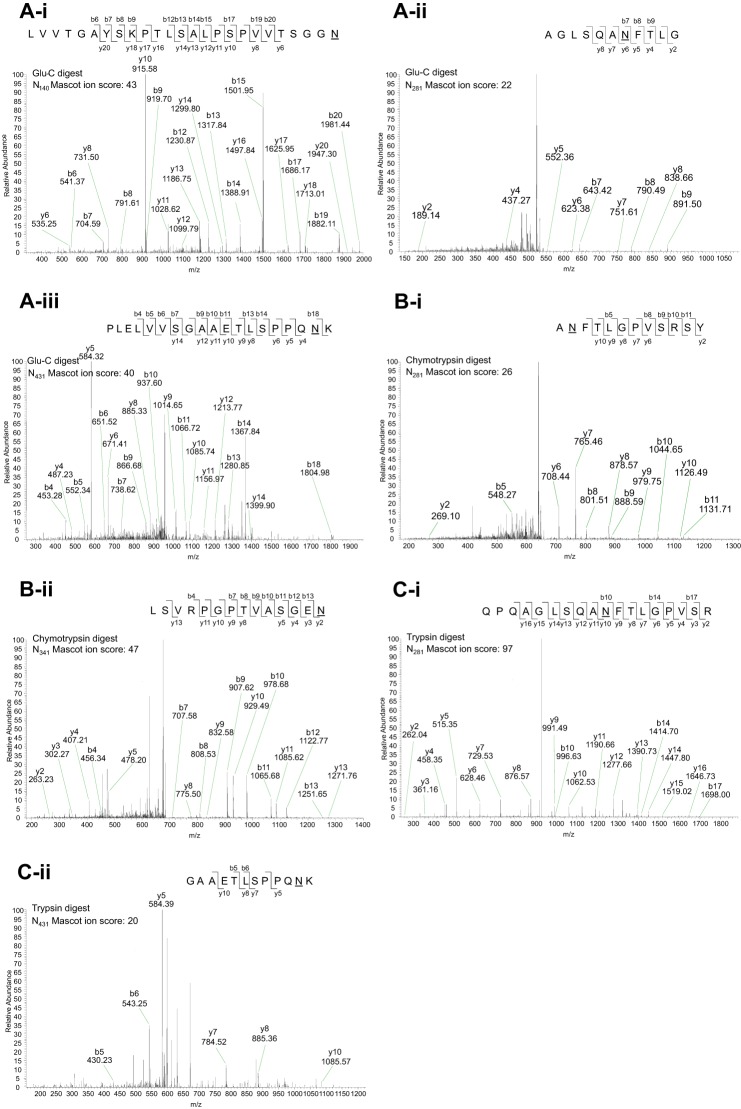
FIGURE 3.
Representative nano-LC-MS/MS of PNGase F-deglycosylated tryptic-digested peptides of mammalian rLILRA3 confirmed four predicted N-glycosylation sites. A, in-gel peptide digestion of deglycosylated rLILRA3 with Glu-C showed deamidation of asparagine to aspartic acid at Asn140 (panel i), Asn281 (panel ii), and Asn431 (panel iii) indicating N-linked glycosylation of these sites. B, digestion with chymotrypsin showed deamidation at Asn281 (panel i) and Asn341 (panel ii). C, digestion with trypsin detected Asn281 (panel i) and Asn431 (panel ii). It is noteworthy that some sites were detected in peptides digested by more than one enzyme, and none of the enzymes provided full peptide coverage. The predicted Asn302 was not detected. The sequence of the peptide, the fragmentation pattern, and the detected fragment ions are shown at the top right in each panel. b ions contain the N-terminal region of the peptide; y ions contain the C-terminal region of the peptide. Deamidation of asparagine to aspartic acid is designated as “N” with an underscore.