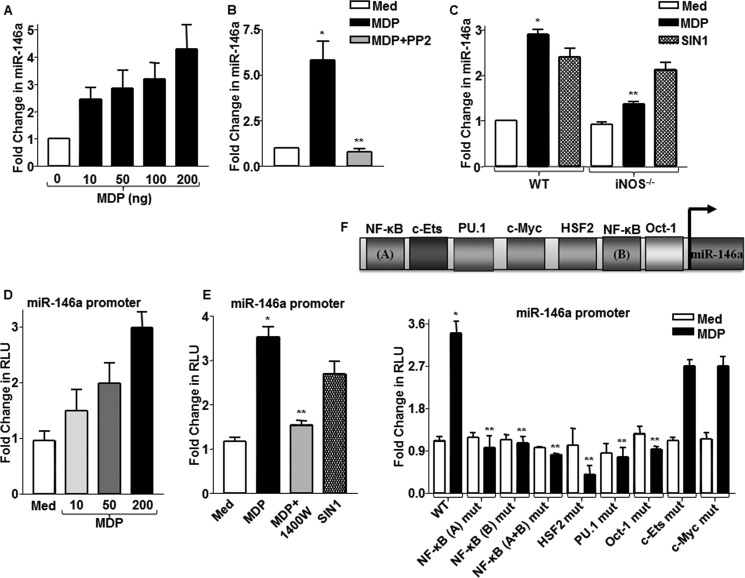
FIGURE 4.
NOD2/NO axis regulates expression of miR-146a. A, miR-146a expression was determined in macrophages stimulated with MDP using quantitative real time RT-PCR. B, macrophages pretreated with RIP2 kinase inhibitor, PP2, were assayed for miR-146a expression after MDP treatment. *, p < 0.05 versus control; **, p < 0.05 versus MDP treatment (one-way ANOVA). Med, medium. C, iNOS null or WT macrophages were treated with MDP or SIN1, and expression levels of miR-146a were analyzed as described. WT, wild type; iNOS−/−, iNOS knock-out. D and E, miR-146a promoter-transfected cells were treated with the indicated concentrations of MDP (D) or with 1400W, iNOS activity inhibitor, prior to MDP stimulation (E), and miR-146a promoter luciferase activity was measured. Alternatively, cells were stimulated with SIN1 in panel E, and miR-146a promoter activity was assayed. *, p < 0.05 versus WT control; **, p < 0.05 versus WT MDP (one-way ANOVA). RLU, relative luciferase unit. F, WT miR-146a promoter luciferase construct or mutant miR-146a promoter luciferase constructs for binding sites of indicated transcription factors were used to assess MDP-induced miR-146a promoter reporter activity by luciferase assay (mean ± S.E., n = 3). *, p < 0.05 versus WT control; **, p < 0.05 versus WT MDP (one-way ANOVA).