FIGURE 6.
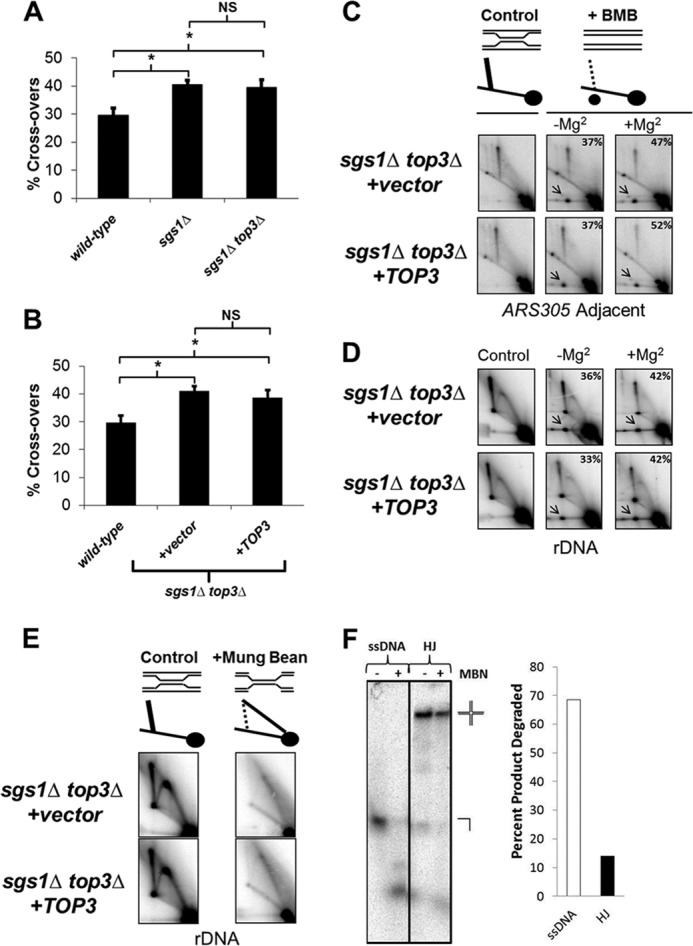
Unassisted Top3 promotes Rec-X resolution but not HJ resolution. A, wild-type, sgs1Δ, and sgs1Δ top3Δ cells were transformed with the ADE2 cassette, and the ratios of crossover-containing to total (crossover + non-crossover) colonies are plotted on the y axis. Data are from three independent experiments and error bars represent the S.E.; *, p < 0.05; NS, not significant. B, identical to A except testing the effects of Top3 overexpression. C and D, schematic: resolved products are indicated by the new spot in the right panel. Southern blot of genomic DNA extracted from sgs1Δ top3Δ cells containing either vector or TOP3 overexpression plasmid, showing resolution of X-shaped molecules into branch migrated products (arrows) at the ARS305 adjacent fragment (C) and rDNA fragment (D). Samples were incubated at 65 °C in branch migration buffer (BMB) ± Mg2+ between the first and second dimensions of electrophoresis. Percentages of branch migrated products are indicated in the top right corner of panels. E, MBN treatment of genomic DNA extracted from sgs1Δ top3Δ cells containing either vector or TOP3 overexpression plasmid, showing alteration in rDNA migration, consistent with Rec-X structures as indicated in the schematic. F, 8% native polyacrylamide gel showing MBN digestion products of a 5′-32P-end labeled 50-base long oligonucleotide alone (ssDNA) or annealed into a synthetic HJ substrate that was added into genomic DNA MBN digests under the same conditions as in E. The percentages of substrates digested are shown on the right. Note that the samples for C and E are from cells grown for 1.5 h in 0.033% MMS, prior to Top3-induced differences in X-structure resolution, whereas samples for D are from cells 2 h after release from MMS.
