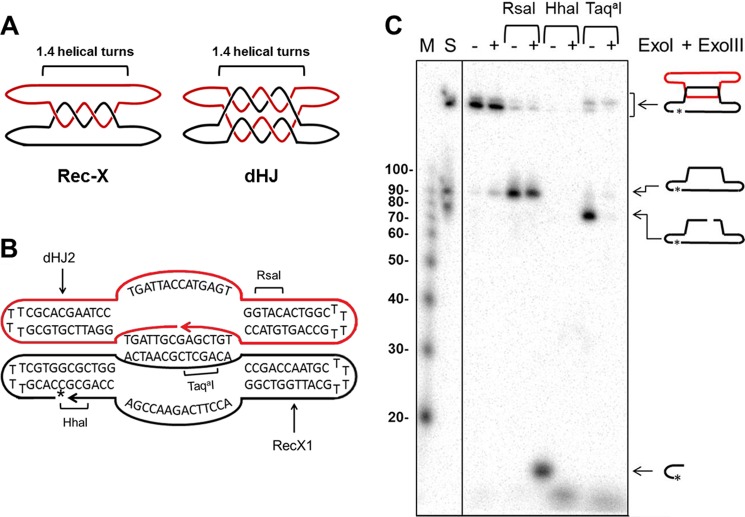
FIGURE 7.
Characterization of synthetic Rec-X structure. A, schematic representations of the Rec-X and dHJ substrates, showing where the two oligonucleotides are topologically interlinked. For simplicity, the helical turns are not drawn in subsequent figures. B, Rec-X map showing the sequences of the RecX1 and dHJ2 oligos that compose it, regions of double-stranded DNA, and RsaI, HhaI, and TaqaI cut sites. The 32P-labeled phosphate is indicated by an asterisk. C, 8% polyacrylamide denaturing gel confirming the structure of Rec-X. First lane (M), 5′-32P-end labeled 10-bp DNA Step ladder (Promega); second lane 2 (S) standard comprising Rec-X, and ligated and unligated forms of the 80-nt long RecX1 oligo; third and fourth lanes, Rec-X without restriction enzyme treatment; fifth and sixth lanes, digested with RsaI; seventh and eighth lanes, digested with HhaI; ninth and tenth lanes, digested with TaqaI. Fourth, sixth, eighth, and tenth lanes, samples were further treated with E. coli ExoI and ExoIII, which degrade single-stranded and double-stranded DNA, respectively, in a 3′ → 5′ direction. Note that the TaqaI digest was performed at 47 °C, indicating that the inter-strand duplex region remained at least partially double stranded during the Top3 decatenation experiments, even at an elevated temperature. In addition we independently confirmed the structure of dHJ in a fashion similar to the characterization of the Rec-X (data not shown), as described previously (43).