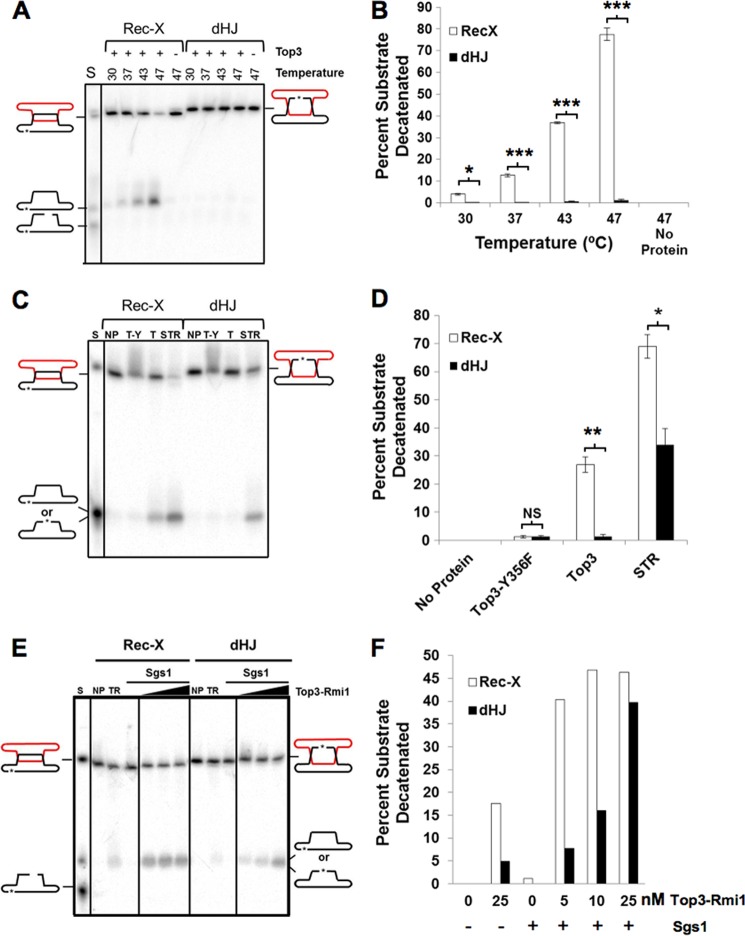
FIGURE 8.
Top3 decatenates a Rec-X but not a dHJ substrate. A, C, and E, samples were run on denaturing polyacrylamide gels. A, decatenation of Rec-X but not dHJ by Top3. DNA substrates were incubated alone or with 40 nm Top3 at the indicated temperatures. The products of Top3-mediated decatenation migrate with the closed circular form. C, decatenation of a Rec-X by Top3 alone, but not by catalytically inactive Top3, and decatenation of a Rec-X and a dHJ by the STR complex. DNA substrates were incubated alone (NP), with 40 nm Top3-Y356F (T-Y) or Top3 (T), or 40 nm Top3-Rmi1 together with 0.6 nm Sgs1 (STR) at 37 °C. E, decatenation of a Rec-X and a dHJ by the STR complex. DNA substrates were incubated alone (NP), or with 1.2 nm Sgs1 and 0, 5, 10, or 25 nm Top3-Rmi1. B, D, and F, quantification of the data in A–C, respectively. The mobility standard (S) for all gels was made by mixing equal amounts of duplex substrate (Rec-X or dHJ), and circular and linear forms of the labeled strand. Percentages were corrected for background by subtracting the value of the no protein control for each substrate (n = 3 for B and D), *, p < 0.05; **, p < 0.01; ***, p < 0.0002.