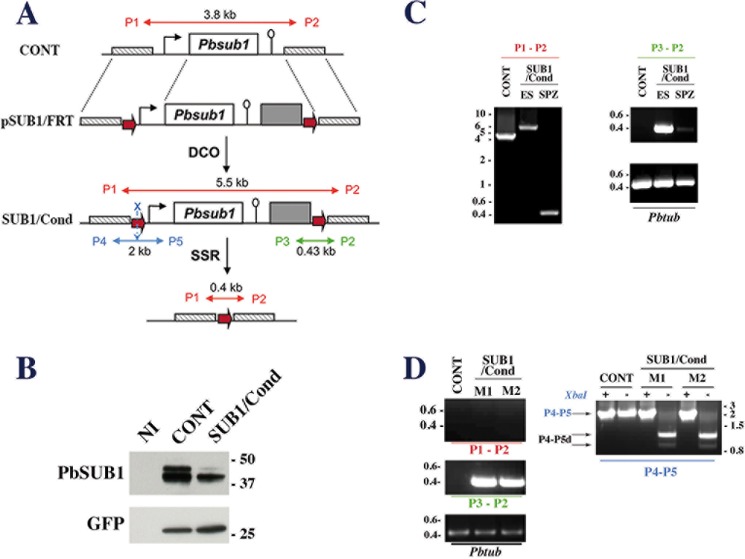
FIGURE 2.
Generation of the P. berghei conditional SUB1/Cond mutant. A, schematic representations of the wild-type Pbsub1 locus in control (CONT) parental parasites, the targeting plasmid pSUB1/FRT (top), the expected Pbsub1 recombinant locus in the SUB1/Cond clone obtained following a double crossover event (DCO; middle), and the recombinant Pbsub1-null locus after the FLP-recombinase mediated site-specific recombination (SSR; bottom). The linearized pSUB1/FRT plasmid carries the following from 5′ to 3′: a 736-bp fragment located upstream of the sub1 locus (cross-hatched box), an FRT site (red arrow), the Pbsub1 locus composed of a 5′-untranslated regulatory sequence (1177 bp; black arrow), the Pbsub1 coding sequence (1799 bp; open box), a 3′-untranslated regulatory sequence (503 bp; open lollipop), the human dihydrofolate reductase cassette (gray box), the second FRT site, and a 673-bp fragment located downstream of the Pbsub1 locus. The P1 and P2 primers amplify a 3.8- or 5.5-kb fragment on CONT or SUB1/Cond loci, respectively, and a 0.4-kb fragment following FLP-specific SSR excision of Pbsub1. P3/P2 and P4/P5 couples of primers are specific for the SUB1/Cond recombinant locus; P3 and P2 amplify a 0.43-kb fragment, whereas P4 and P5 amplify a 2-kb fragment surrounding the 5′ FRT site, which contains an XbaI restriction site (blue X). B, Western blot analysis of SUB1/Cond clone5 and CONT erythrocytic segmented schizonts probed for PbSUB1. Protein extracts corresponding to 5 × 105 infected red blood cells and an equal number of non-infected erythrocytes (NI) were probed by 1D11 anti-PbSUB1 and anti-GFP antibodies; the latter was used as a loading control. C, PCR analysis of the Pbsub1 locus of the recombinant SUB1/Cond clone 5 compared with the parental (CONT) parasites. P3-P2 and P1-P2 primers were used to analyze the SUB1/Cond gDNA prepared from erythrocytic stages (ES) and sporozoites (SPZ) collected on day 20 postinfection. Primers P1-P2 amplify both the excised (SPZ; 0.4 kb) and non-excised Pbsub1 loci (ES; 5.5 kb) from SUB1/Cond and the parental Pbsub1 locus (CONT; 3.8 kb). Primers P3-P2 amplify only non-excised Pbsub1 locus (0.43 kb) from gDNA prepared from sporozoites and erythrocytic stages of SUB1/Cond parasites. The P. berghei tubulin coding gene (Pbtub) was amplified as a control of gDNA. D, PCR analysis of genomic DNA of CONT and SUB1/Cond erythrocytic stages following infection of mice by sporozoites. Genomic DNA was prepared from the two parasitized mice (M1 and M2) corresponding to experiment 6 (see Table 1) and analyzed using P1-P2, P3-P2, and P4-P5 primers as indicated in C. As shown by the amplification of a 0.43-kb fragment with the P3-P4 primers and a 2-kb fragment with the P4-P5 primers, erythrocytic parasites collected from both mice display a SUB1/Cond genotype. XbaI-digested (+) or undigested (−) P4-P5 fragments (P4-P5, blue characters) are shown. In the presence of a complete 5′ FRT site containing the XbaI site, two fragments of 1.2 and 0.8 kb are generated (P4-P5d; black arrows) and are the signature of a complete SUB1/Cond recombinant locus. Importantly, no fragment of 0.4 kb corresponding to a Pbsub1 excised locus could be amplified with P1-P2 primers. Molecular mass markers are in kilobase pairs and in kilodaltons.