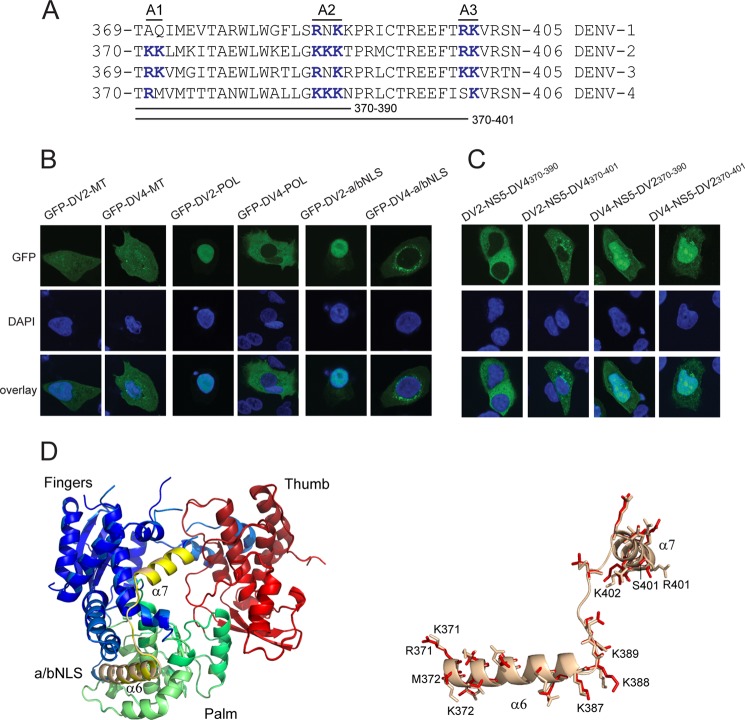
FIGURE 4.
DENV-4 NS5 does not have a functional a/bNLS. A, alignment of the DENV-1, -2, -3, and 4 NS5 amino acid sequences corresponding to the DENV-2 NS5 a/bNLS. Three basic amino acid clusters found in the DENV-2 a/bNLS (termed A1, A2, and A3) and conserved basic amino acid sequences in the other sequences are shown in boldface type and colored blue. The amino acid sequences interchanged between DENV-2 and -4 NS5 are shown. B, CLSM images of A549 cells transfected with pcGFP-DV2-MT (GFP-DV2-MT) and pcGFP-DV4-MT (GFP-DV4-MT) (left panels), pcGFP-DV2-POL (GFP-DV2-POL) and pcGFP-DV4-POL (GFP-DV4-POL) (middle panels), and pc2xGFP-DV2-NLS (GFP-DV2-a/bNLS) and pc2xGFP-DV4-NLS (GFP-DV4-a/bNLS) (right panels). C, CLSM images of A549 cells transfected with pcGFP-DV2-NS5-DV4(370–390) (DV2-NS5-DV4370–390), pcGFP-DV2-NS5-DV4(370–401) (DV2-NS5-DV4370–401), pcGFP-DV4-NS5-DV2(370–390) (DV4-NS5-DV2370–390), and pcGFP-DV4-NS5(370–401) (DV4-NS5-DV2370–401). The transfected cells were analyzed for GFP fluorescence at 24 h post-transfection. Nuclear DNA was stained with DAPI in all experiments. D, superimposition of models of the POL structures of DENV-2 NS5 and DENV-2 NS5 containing DENV-4 amino acids 370–401. The DENV-2 POL sequences were used to generate hidden Markov models (72), which were then compared against a database of hidden Markov models with solved atomic structures. The MODELLER software suite (73) was used to generate the three-dimensional models based upon the x-ray structure of the DENV-3 NS5 POL domain (60). PyMOL (74) was the used to align the structures and for molecular graphics. The aligned POL structures are shown on the left, with the fingers, thumb, and palm subdomains and the a/bNLS (containing α helices 6 and 7) shaded in blue/light blue, red/salmon, green/pale green, and yellow/sand for the DENV-2 and DENV-2/4 POLs, respectively. On the right, an expanded view of the a/bNLS alignment is shown in schematic form. Amino acids that differ in side chain residue or orientation are shown in stick form. The amino acids in the A1, A2, and A3 clusters are numbered for the DENV-2 and -4 sequences.