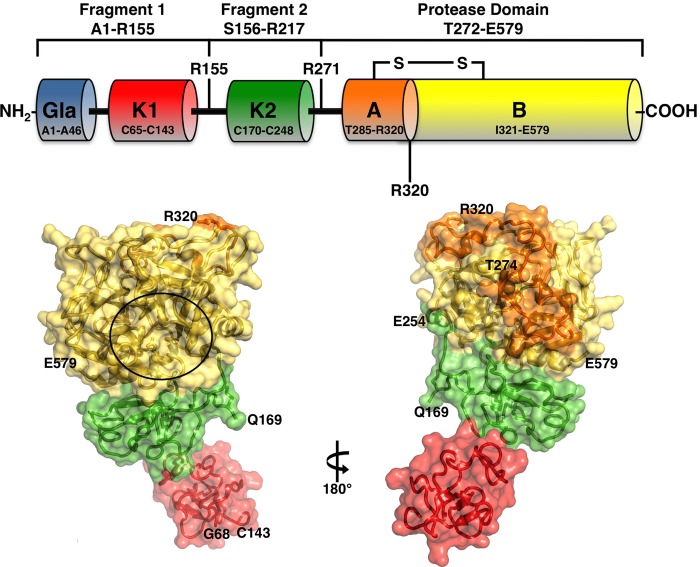
FIGURE 1.
Top, schematic representation of prothrombin composed of fragment 1 (residues 1–155), fragment 2 (residues 156–271), and a protease domain (residues 272–579). Fragment-1 contains a Gla domain (residues 1–44) and a kringle (residues 65–143), fragment-2 contains a second kringle (residues 170–248), and the protease domain contains the A (residues 272–320) and B (residues 321–579) chains. Thrombin is generated by two cleavages at Arg-271 and Arg-320, producing the inactive precursor prethrombin-2 and the active intermediate meizothrombin, respectively. The A and B chain remain covalently attached after activation through the disulfide bond Cys-293–Cys-439 (Cys-1–Cys-122). Cleavage at Arg-284 by thrombin itself reduces the length of the A chain to its final 36 amino acids composition. Bottom, x-ray crystal structure of Gla-domainless prothrombin with kringle-1 (red) positioned at an angle of 36° relative to kringle 2 (green) that is coaxial to the protease domain (B chain in yellow and A chain in orange). The active site region is indicated by a circle, and the termini for each domain visible in the orientation are noted. Of the two sites of cleavage, Arg-320 (Arg-15) in the activation domain is visible, but Arg-271 is part of a 20-residue segment missing in the electron density map because of disorder.