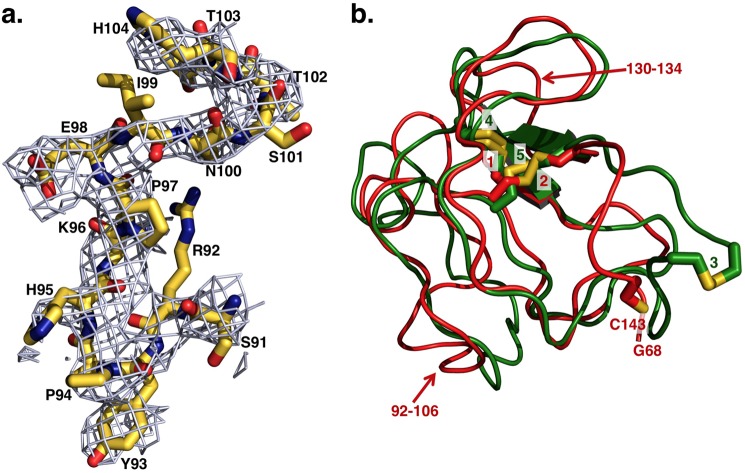
FIGURE 2.
a, representative extra electron density detected for fragment 1 after molecular replacement and before refinement. The extra electron density shown as 2Fo − Fc map contoured at 1 σ enables assignment of residues of fragment 1 of Gla-domainless prothrombin with confidence. Shown as sticks are residues Ser-91–His-104 of kringle-1 with the distinguishing Pro-94 that is the single Pro in the cis conformation in the entire kringle, as first identified in the high resolution structure of bovine kringle-1 (6). These residues refer to the structure after refinement, deposited in the Protein Data Bank as entry 4HZH, and show the quality of the model built on the extra electron density detected before refinement after initial molecular replacement. b, overlay of kringle-1 (red) and kringle-2 (green) of Gla-domainless prothrombin reveals the basic similarity between the two domains. Because of the absence of Cys-65 in the electron density map, Cys-143 is unpaired, and only two disulfide bonds (1 = Cys-86–Cys-125, 2 = Cys-114–Cys-138) are detected in kringle-1. All three disulfide bonds (3 = Cys-170–Cys-248, 4 = Cys-191–Cys-231, 5 = Cys-219–Cys-243) are detected for kringle-2. The two kringles align with an r.m.s.d. = 1.2 Å, and the most notable differences (noted by arrows) are a helical segment in kringle-2 replaced by an unstructured coil (residues 92–106) in kringle-1 (see also panel a) and residues 130–134 of kringle-1 shifted relative to the homologous region in kringle-2 by ∼3 Å.