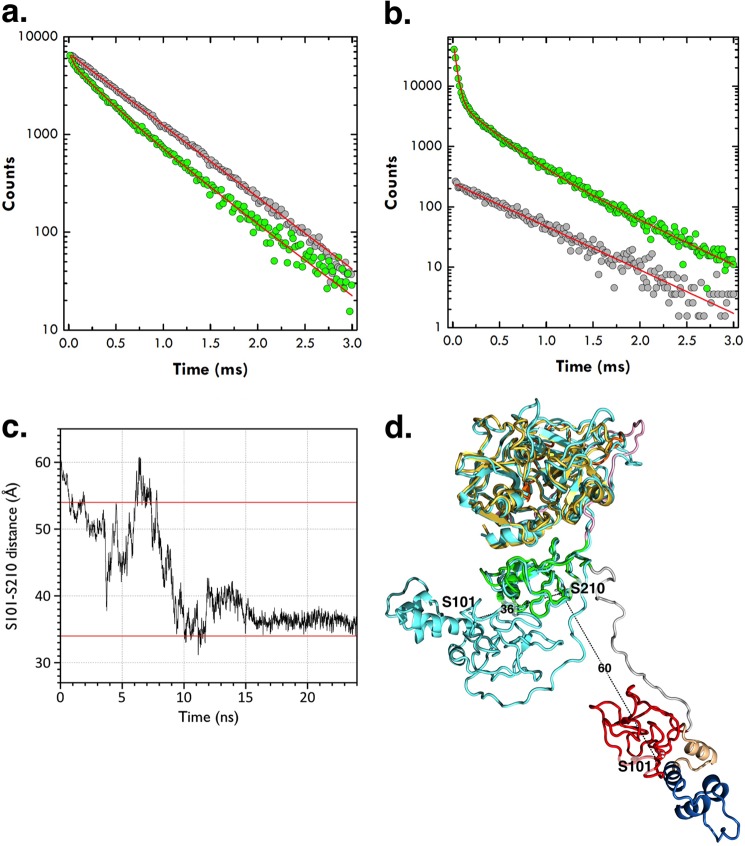
FIGURE 3.
a and b, semilog plot of lifetime data for the LRET donor-acceptor pair conjugated to residues 101 in kringle-1 and 210 in kringle-2 of the full-length prothrombin mutant S101C/S210C. The donor quenching (a, green circles) and acceptor sensitization (b, green circles) both obey the same triple-exponential decay comprising a short lifetime of 33.8 ± 0.5 μs with almost complete (94%) energy transfer and a population of 40% and a long lifetime of 272 ± 4 μs with 55% energy transfer and a population of 60%. The third, slowest lifetime (606 ± 9 μs) is identical to the decay of the donor-only control and free Eu3+ chelate (gray circles). The donor-acceptor distances associated with the slow (272 ± 4 μs) and fast (33.8 ± 0.5 μs) decays derived from the Förster equation (see “Experimental Procedures”) are 54 ± 2 and ≤34 Å, respectively. c, molecular dynamics simulations of the prothrombin conformation shown as the Cα-Cα distance between Ser-101 in kringle-1 and Ser-210 in kringle-2 as a function of time, over a 24-ns trajectory. Horizontal red lines depict the values of 54 and 34 Å determined by LRET measurements using probes attached to residues 101 and 210 mutated to Cys. d, overlay of the starting (with domains colored as in Fig. 1) and ending (cyan) prothrombin structures from the molecular dynamics simulation (see panel c). The starting structure is the model of prothrombin bound to prothrombinase (14), and the ending structure is the average of the last 5 ns of the simulation. Residues Ser-101 and Ser-210 are rendered as sticks with Cα-Cα distances indicated in Å.