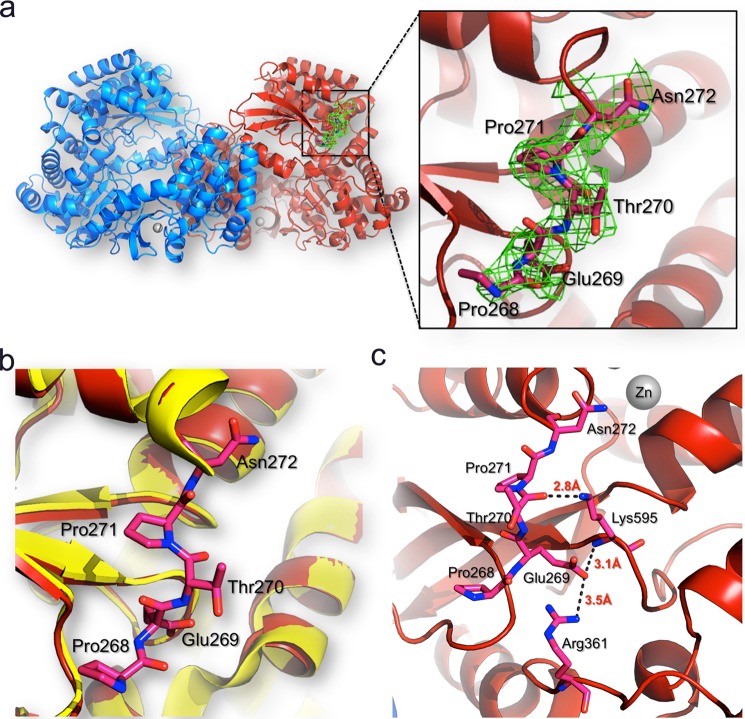
FIGURE 6.
Interactions of residues 268–271 with the DENV3 RdRp protein core. a, location of residues 268–271 (represented as sticks and colored in pink) in the RdRp dimer structure. The two protein monomers are represented as blue and red ribbons, respectively. For clarity, the linker is shown only for one monomer but has the same conformation in the other monomer. Inset, residues 268–271 are visible in the refined DENV-3 RdRp265–900 structure (this work). The 2Fo − Fc electron density map (green) calculated with phases from the refined model and displayed at 1σ level is overlaid on the current model for linker residues. b, the RdRp272–900 structure (Protein Data Bank code 2J7U; in yellow) is overlaid on the RdRp265–900 structure (Protein Data Bank code 4C11; in red), and linker residues are displayed as red sticks and labeled. c, atomic interactions established between the linker residues with the core domain of the polymerase. Hydrogen bonds between linker residues and the core polymerase domain are represented by dashes with the respective distances indicated.