Abstract
Protein ubiquitination occurs through formation of an isopeptide bond between the C-terminal glycine of ubiquitin (Ub) and the ɛ-amino group of a substrate lysine residue. This post-translational modification, which occurs through the attachment of single and/or multiple copies of mono-ubiquitin and poly-ubiquitin chains, is involved in crucial cellular events such as protein degradation, cell-cycle regulation and DNA repair. The abnormal functioning of ubiquitin pathways is also implicated in the pathogenesis of several human diseases ranging from cancer to neurodegeneration. However, despite the undoubted biological importance, understanding the molecular basis of how ubiquitination regulates different pathways has up to now been strongly limited by the difficulty of producing the amounts of highly homogeneous samples that are needed for a structural characterization by X-ray crystallography and/or NMR. Here, we report on the production of milligrams of highly pure Josephin mono-ubiquitinated on lysine 117 through large scale in vitro enzymatic ubiquitination. Josephin is the catalytic domain of ataxin-3, a protein responsible for spinocerebellar ataxia type 3. Ataxin-3 is the first deubiquitinating enzyme (DUB) reported to be activated by mono-ubiquitination. We demonstrate that the samples produced with the described method are correctly folded and suitable for structural studies. The protocol allows facile selective labelling of the components. Our results provide an important proof-of-concept that may pave the way to new approaches to the in vitro study of ubiquitinated proteins.
Keywords: Ubiquitin, Post-translational modification, Isopeptide bond, Josephin, Spinocerebellar ataxia type 3, Machado–Joseph disease, Deubiquitinating enzyme
Abbreviations: ATP, adenosine triphosphate; DTT, dithiothreitol; DUB, deubiquitinating enzyme; GST, glutathione-S-transferase; HSQC, heteronuclear single quantum coherence; IAA, iodoacetamide; JosK117-only, Josephin mutant in which all lysines but K117 are mutated; MS/MS tandem, mass spectrometry; NMR, nuclear magnetic resonance; PDB, Protein Data Bank; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; Tris–HCl, 2-amino-2-(hydroxymethyl)-1,3-propanediol hydrochloride
Graphical abstract
Highlights
-
•
We set up a protocol for large-scale in vitro enzymatic ubiqitination.
-
•
This produced milligrams of highly pure mono-ubiquitinated Josephin domain of ataxin-3.
-
•
We applied an alternative labelling scheme for the structural characterization of the sample by NMR.
-
•
Ubiquitin covalently linked on lysine 117 directly interacts with Josephin but does not alter the overall fold of the protein.
1. Introduction
Post-translational modifications of proteins regulate a wide variety of cellular events [1]. One the most important of such modifications is ubiquitination, a reaction first described ∼40 years ago [2]. It consists of the covalent attachment of the ɛ-amino group of a target protein lysine to the carboxylic group of ubiquitin C-terminal glycine via an isopeptide bond. In vivo, ubiquitin conjugation is performed by a cascade of three classes of enzymes, named ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3), and is reversed by deubiquitinating enzymes (DUBs). Proteins can be mono-ubiquitinated, multi-monoubiquitinated, or poly-ubiquitinated [3,4]. Initially, ubiquitin was described as a molecular death-tag, promoting protein degradation by the proteasome [5,6]. More recently, several other roles of ubiquitin have been discovered, broadening the field of action of the protein to cellular processes such as signalling, cell cycle regulation, and DNA repair [7–9]. Mono-ubiquitination can also have different functional consequences for a target protein, such as changes in binding properties, subcellular localization and activity [10].
Despite its fundamental biological role, very little is known about how ubiquitination influences the structure/functions of the covalently linked cargo proteins. Yet, unlike other less invasive modifications such as phosphorylation, which introduces a relatively small group (80 Da), ubiquitination results in the addition of one or more repeats of a globular protein of 76 amino acids (about 8.5 kDa). The interaction between ubiquitin and the cargo protein could thus produce major conformational changes. The main reason for this lack of knowledge is that the production of ubiquitinated proteins in the amounts required for structural characterization remains challenging [11].
The relatively few structures available in Protein Data Bank (PDB) of proteins bound to ubiquitin correspond either to non-covalent complexes, or to complexes in which the C-terminal glycine of ubiquitin is covalently bound through a thioester bond to a cysteine of a substrate protein. Among this second group are enzymes with a reactive cysteine in the active site having ubiquitin (i.e. E1, E2, and E3 enzymes) or ubiquitin chains (i.e. DUBs) as a substrate. For DUBs in particular, ubiquitin covalent binding is obtained using suicidal irreversible inhibitors as ubiquitin aldheyde or ubiquitin vinyl sulfone [12–18]. The structures of these enzymes correspond to bona fide reaction intermediates, in which the substrate (ubiquitin) is covalently linked to the active site. While appropriate for specific examples, this approach remains highly unsatisfactory to describe the effect of ubiquitination as it significantly alters the structural and geometrical relationship between cargo and ubiquitin.
Different chemical and enzymatic strategies have more recently been proposed to covalently link the ɛ-amino group of a target protein lysine to the carboxylate group of the C-terminal glycine of ubiquitin. Non-enzymatic strategies consist in semi-synthetic methods that often exploit intein chemistry and multiple protection and deprotection steps [19]. Histone H2B and α-synuclein have, for instance, been mono-ubiquitinated using chemical ligation [19,20]. Several chemical methods consist in the formation of isopeptide bond mimics [21]. A new synthetic method named GOLAP could potentially be applied to protein ubiquitination, but it requires protein refolding after ligation [22,23]. A different approach is based on the use of E1, E2 and E3 enzymes to catalyze the formation of a native isopeptide bond. A recent paper suggests the co-expression in Escherichia coli of all the proteins necessary for the ubiquitination cascade [24]. These methods are, however, overall laborious and of not easy implementation.
Here, we show how we can, through the careful setup and optimization of an enzymatic in vitro approach, produce milligrams of a mono-ubiquitinated protein in quantities suitable for structural studies. We used Josephin that is the catalytic domain of ataxin-3, a DUB responsible for spinocerebellar ataxia of type 3 (or Machado–Joseph disease). Josephin is a papain-like cysteine protease that preferentially cleaves long ubiquitin chains [25]. The crystal structure of the Josephin domain of an ataxin-3-like protein covalently attached to ubiquitin through the catalytic cysteine has been published [26]. This complex is representative of how Josephin interacts with its substrate. However, besides binding ubiquitin as a substrate, ataxin-3 can itself be mono-ubiquitinated in the cell with the major site of ubiquitination being lysine 117 on the Josephin domain [27,28]. Mono-ubiquitination results in the increase of the DUB activity of the protein through a still elusive molecular mechanism [27,28]. Understanding how ubiquitination leads to enzyme activation demands the production of properly (through an isopeptide bond) mono-ubiquitinated Josephin. Our goal was therefore to produce a homogeneous sample of Josephin mono-ubiquitinated at lysine 117 in quantities (mg) amenable for structural studies. The strategy that we describe constitutes an important proof-of-concept and a new step towards understanding protein regulation by ubiquitination.
2. Results
2.1. Mono-ubiquitinated Josephin can be produced in vitro enzymatically
The pipeline followed in this paper is summarized in Fig. 1.
Fig. 1.
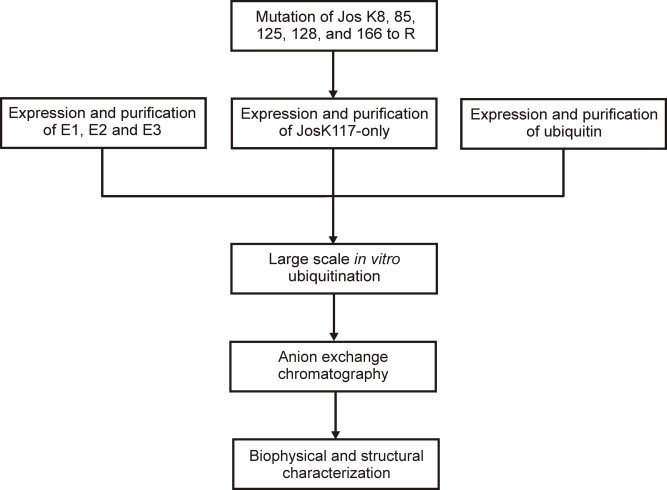
Scheme of the protocol set up for the production of mono-ubiquitinated JosK117-only.
We have previously demonstrated that lysine 117 is the primary site of ubiquitination of Josephin both in vitro and in cells [28]. However, it cannot in principle be ruled out that minor fractions of Josephin are ubiquitinated on a different lysine residue [28]. Since sample homogeneity is an essential prerequisite for structural studies, we used a Josephin mutant (JosK117-only), in which all lysines but K117 (i.e. K8, 85, 125, 128 and 166) were mutated to arginines. These mutations will direct ubiquitination specifically on lysine 117. According to a previously described protocol [27], in vitro ubiquitination of Josephin can be catalyzed by 0.16 μM E1, 8 μM UbcH5a and 1 μM CHIP, 50 μM Ub, 4.5 mM MgCl2, 4.5 mM ATP in buffer 50 mM Tris–HCl, 50 mM KCl, 0.2 mM DTT, pH 7.5 for 2 h at 37 °C. However, our previous studies on Josephin stability demonstrated that the protein is prone to aggregation [30], and that temperature and/or high ionic strength increase the tendency of Josephin to form aggregates. To reduce the risk of protein aggregation, the enzymatic reaction was run at 25 °C for 20 h instead of 37 °C for 2 h and no KCl was added to the ubiquitination buffer. Commercial enzymes were initially used to set up the protocol for large-scale production of mono-ubiquitinated Josephin. Samples from the reaction mixture were collected before addition of ATP (t = 0), after 3 and 20 h. A band at about 30 kDa, corresponding to the molecular weight of JosK117-only (c.a. 21 kDa) covalently linked to ubiquitin (c.a. 8.5 kDa) appeared after 3 h and its intensity increased over time as the mono-ubiquitinated product formed (Fig. 2, lanes 1–3). In parallel, JosK117-only and ubiquitin were consumed. We then increased the concentrations of E1, UbcH5a, and CHIP to ubiquitinate the majority of JosK117-only in the mixture. At the same time, we scaled up Josephin and ubiquitin concentrations from 12 μM and 50 μM to 50 μM and 250 μM, respectively (Fig. 2, lanes 4–6). Using these conditions, we obtained a satisfactory yield of mono-ubiquitinated Josephin.
Fig. 2.
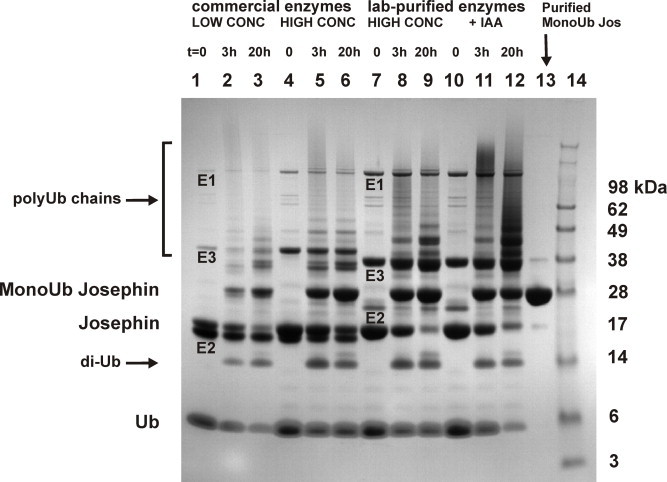
In vitro ubiquitination of JosK117-only. All reactions are monitored at t = 0, 3 and 20 h. Lanes 1–3: reaction using commercial enzymes (0.16 μM E1, 8 μM UbcH5a and 1 μM CHIP), 50 μM ubiquitin and 12 μM JosK117-only at room temperature [27]. Lanes 4–6: reaction using commercial enzymes (1 μM E1, 8 μM UbcH5a, 8 μM CHIP), 250 μM ubiquitin and 50 μM JosK117-only. Lanes 7–9: reaction using enzymes prepared in the lab at the final concentrations as in lanes 4–6. Lanes 10–12: reaction using Josephin pre-treated with IAA. Lane 13: purified mono-ubiquitinated JosK117-only. Lane 14: molecular weight marker. Di-ubiquitin and poly-ubiquitin chains are indicated with arrows. The bands of E1, E2 and E3 enzymes are indicated with annotation below each corresponding band. The Addgene clone of UbcH5a used for the production in the lab comprise an initial sequence deriving from cloning which explains the higher MW with respect to the commercial enzyme.
2.2. Scaling-up the sample production
The formation of the mono-ubiquitinated product further increased using enzymes purified in the laboratory (Fig. 2, lanes 7–9). Since we could produce large amounts of enzymes, we were able to work with larger total volumes of the mixture, which ranged between 5 and 15 ml. These amounts would be prohibitively expensive when using commercial enzymes. The yield of mono-ubiquitinated product remained comparable to that obtained in the small-scale trials. Mono-ubiquitinated JosK117-only is the most abundant product of ubiquitination with the highest yields obtained after overnight reaction (Fig. 2, lane 9). However, we observed also other side-products which form along with mono-ubiquitinated JosK117-only: unanchored poly-ubiquitin chains and poly-ubiquitinated Josephin species. Unanchored ubiquitin chains form by the catalytic action of E1 and UbcH5a since UbcH5a is a promiscuous enzyme which forms K48, K63 and K11 linkages [31]. The formation of di-ubiquitin can be followed on the gel, corresponding to a band with apparent molecular weight of 14 kDa (Fig. 2, bottom arrow). Longer ubiquitin chains gave a characteristic smear band visible in the SDS–PAGE gel (Fig. 2, top arrow).
Interestingly, the overall intensity of the smear band is lower after 20 h compared to 3 h, as Josephin exploits its deubiquitinating activity (Fig. 2, lanes 8 and 9). This could be explained by considering that Josephin is itself a DUB that is able to break both K48 and K63 linkages in poly-ubiquitin chains [32]. To test this hypothesis, we blocked the catalytic cysteine on Josephin with iodoacetamide (IAA), removed the unreacted IAA and ran a parallel ubiquitination reaction (Fig. 2, lanes 10–12). We observed that Josephin inactivation causes a shift of the smear band towards higher molecular weights, consistent with these species not being cleaved by the catalytic inactive Josephin. Ubiquitin is consumed faster and the overall amount of mono-ubiquitinated product is reduced. This result shows that the yield of in vitro ubiquitinated Josephin is influenced by its DUB activity and suggests that Josephin removes the ubiquitin chains (but not the first ubiquitin moiety) attached to lysine 117. It is also likely that Josephin increases the amount of substrate available for the ubiquitination since it breaks the ubiquitin chains and releases mono-ubiquitin in the mixture.
Other “unwanted” products of the enzymatic reaction are formed by elongation of the ubiquitin chain on Josephin by UbcH5a/CHIP. We clearly observed the formation of products with molecular weight of about 39 kDa (Josephin + 2 Ub, band overlapped with CHIP) and 47 kDa (Josephin + 3 Ub) although longer products also form in smaller amounts. To further increase the yield of mono-ubiquitinated Josephin, we attempted to block chain elongation using either methylated ubiquitin (Enzo Lifescience) or an E2 mutant, which cannot extend ubiquitin chains [27]. In both cases we obtained very low yields of the mono-ubiquitinated product (data not shown).
2.3. Purification of mono-ubiquitinated Josephin
As an alternative to the strategy outlined above, we developed a protocol for the separation of mono-ubiquitinated JosK117-only from the poly-ubiquitinated products. Mono-ubiquitinated JosK117-only was separated from poly-ubiquitinated products, unreacted substrates and ubiquitination enzymes by anion exchange chromatography (Fig. 3). Initially all components of the reaction mixture are bound to the column (including unreacted ATP and AMP produced by catalysis). The presence of these nucleotides produces a strong UV signal, which saturates the absorbance detector and covers protein peaks at 280 nm. ATP and AMP were conveniently separated from the proteins directly on the column using a wash step at 0.1 M NaCl.
Fig. 3.
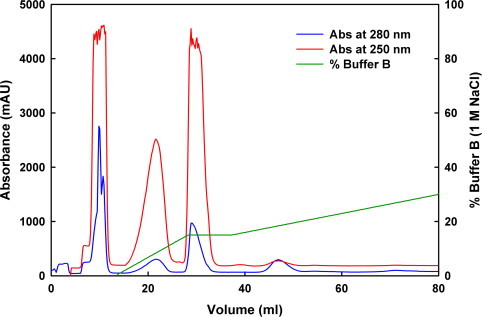
Anion exchange purification of mono-ubiquitinated JosK117-only. Absorbance at 280 nm (blue), 250 nm (red) and percentage of buffer B (1 M NaCl) are reported. Mono-ubiquitinated JosK117-only elutes at ∼47 ml. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A salt gradient was then applied to purify mono-ubiquitinated JosK117-only. E1, UbcH5a, CHIP, unreacted ubiquitin, and ubiquitin chains were easily removed as they elute at different NaCl concentrations with respect to all Josephin species. Mono-ubiquitinated JosK117-only could be efficiently separated from unreacted and poly-ubiquitinated Josephin species although the theoretical isoelectric points are quite close (JosK117-only has a pI of 4.7 while that of mono-ubiquitinated JosK117-only is 4.9). We applied a shallow salt gradient, typically 0.1–0.3 M NaCl in a total buffer volume of about 300 ml. This method yielded to a high purity product as confirmed by SDS–PAGE (Fig. 2, lane 13). Mass spectrometry analysis after trypsin digestion was performed to check the isopeptide bond formation. Trypsin cleaves ubiquitin covalently-linked to a protein at the junction between arginine 74 and glycine 75, thus producing a GG signature. A peptide with mass corresponding to residues 111–124 from JosK117-only plus the two C-terminal glycines from ubiquitin was isolated (Fig. 4, top, right arrow). The data also comprise a peptide with mass corresponding to the first 10 amino acids of JosK117-only (Fig. 4, top, left arrow), but no peak with the same mass plus the GG ubiquitin signature was found (expected mass of 1304 Da), excluding ubiquitination at the N-terminus. Fragmentation by MS/MS of the peptide 111–124 confirmed that K117 is linked via an isopeptide bond to glycine 76 of ubiquitin (Fig. 4, bottom).
Fig. 4.
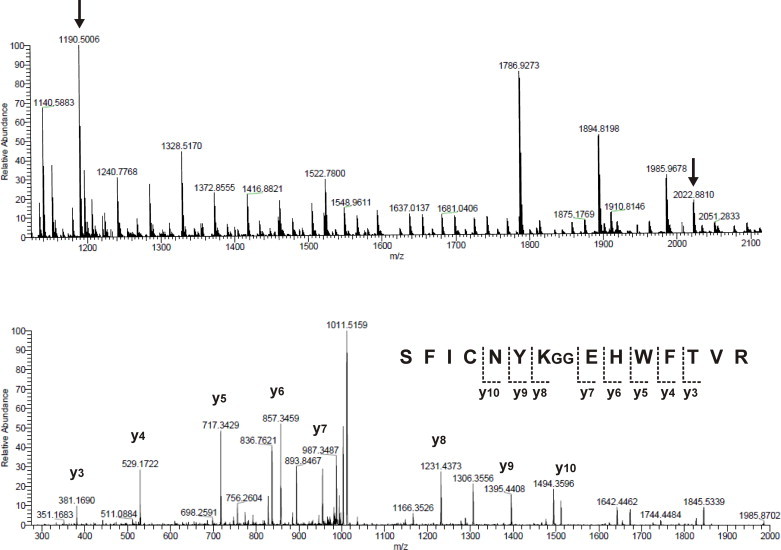
MS analysis of JosK117-only. Top: MS spectrum of mono-ubiquitinated JosK117-only after trypsin digestion. The left arrow corresponds to Josephin GAMESIFHER N-terminal peptide, the right arrow indicates the mass of amino acids 111–124 including the ubiquitin C-terminal GG dipeptide covalently attached to lysine 117. Bottom: fragmentation with MS/MS of the peptide containing lysine 117.
2.4. Josephin mono-ubiquitination does not affect the fold
To characterize the obtained sample, we compared the 1H–15N HSQC spectrum of JosK117-only and Josephin wild-type to check the effect of the introduced mutations (five lysines mutated to arginines). The chemical shift variations are modest demonstrating that JosK117-only has a very similar fold as compared to the wild-type protein (Fig. 5A). This proves that the conservative mutations introduced to specifically direct ubiquitin covalent binding on lysine 117 do not alter the structure of the protein and that the mutant JosK117-only is an appropriate model to study Josephin ubiquitination.
Fig. 5.
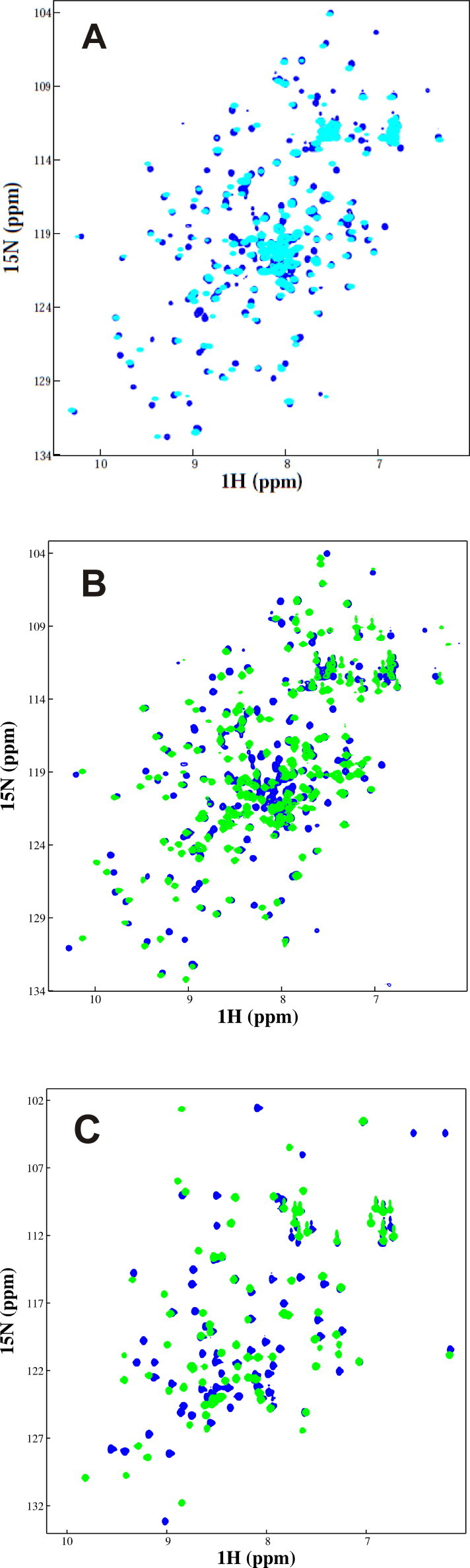
Comparison of NMR spectra of different Josephin samples. (A) 15N HSQC spectrum of labelled Josephin wild-type (in cyan) superimposed to the spectrum of labelled JosK117-only (in blue). (B) 15N HSQC spectrum of labelled JosK117-only covalently linked to unlabelled ubiquitin (in green) superimposed to the spectrum of labelled JosK117-only (in blue). (C) 15N HSQC spectrum of labelled ubiquitin covalently linked to unlabelled JosK117-only (in green) superimposed to the spectrum of labelled ubiquitin (in blue).The samples contain 300 μM proteins in 20 mM Na phosphate pH 6.5, 2 mM DTT. The spectra were recorded at 700 MHz and 25 °C. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The protocol outlined here allows the facile selective labelling of the specific moieties. We can for instance produce 15N labelled JosK117-only attached to unlabelled ubiquitin (Fig. 5B) or 15N labelled ubiquitin attached to unlabelled JosK117-only (Fig. 5C).
The 1H–15N HSQC spectrum of mono-ubiquitinated Josephin prepared using 15N JosK117-only and unlabelled ubiquitin was then compared to the spectrum of unmodified 15N JosK117-only (Fig. 5B). Peak dispersion is similar in the two samples, confirming that mono-ubiquitinated JosK117-only is well folded and excluding substantial aggregation. Several chemical shift variations are observed with respect to the non-ubiquitinated protein but, overall, the chemical shift values do not change after ubiquitination for a good percentage of the residues. Likewise, the spectrum of labelled ubiquitin varies in the ubiquitinated sample for some specific residues but without major overall perturbations (Fig. 5C). These data confirm that ubiquitin on lysine 117 directly interacts with Josephin, but the interaction does not have dramatic effects on the overall structure of the protein.
2.5. Discussion
Here, we have described a method for the enzymatic production of highly pure mono-ubiquitinated Josephin with a naturally occurring isopeptide bond. We report a protocol for a large-scale (mg) in vitro enzymatic production (using appropriate E1, E2 and E3 enzymes) of a protein mono-ubiquitinated at a physiological site with a native isopeptide bond. Our data should be compared with recent publications that also describe large-scale in vitro ubiquitination for the production of samples for structural studies. A recent paper reported the structure of a RING E3 ligase, RNF4, in association with UbcH5a linked to ubiquitin [33]. The authors attached ubiquitin to the catalytic site of UbcH5a by insertion of a mutation C85K, where the catalytic cysteine was mutated to lysine and an isopeptide bond was formed in the presence of E1 and ATP. Although the approach is in principle similar to ours, the product did not represent a protein mono-ubiquitinated on a lysine that is commonly ubiquitinated in vivo, but can rather be classified as an E2 reaction intermediate. Another publication reported an NMR characterization of mono-ubiquitinated Ras [34]. The sample was obtained by mutating the lysine known to be ubiquitinated in vivo to cysteine (Ras K147C), and introducing a cysteine at the C-terminus of ubiquitin (G76C mutant). The disulfide bond formed by oxidation of the cysteines mimics the native isopeptide bond but it cannot faithfully reproduce its chemical nature and geometry. Interestingly, no effect on the HSQC spectrum was observed after this covalent binding (S–S) of ubiquitin. A similar approach (disulfide-mediated conjugation) was also used for mono-ubiquitinated PCNA [35], by exploiting intein synthesis and chemical ligation. Mono-ubiquitinated PCNA was produced in another study by splitting the sequence in two auto-assembling fragments at the level of the lysine found to be ubiquitinated in vivo, and expressing ubiquitin together with the first half of the protein [36]. The authors used two glycine residues as a linker in order to mimic the isopeptide bond. While certainly interesting, these approaches might substantially alter the nature of the linkage by changing its length and geometrical/chemical properties. Recently, PCNA mono-ubiquitination was performed via enzymatic reaction in order to obtain a sample for crystallization [37]. This is so far the only structure deposited in PDB of a protein in which ubiquitin is linked by a native isopeptide bond to a specific lysine reported to be ubiquitinated in vivo. This work proves that the enzymatic approach we report is suitable for the structural characterization of mono-ubiquitinated proteins.
The difficulty of producing samples of ubiquitinated proteins for structural characterization has been a serious limitation to understanding yet unexplored allosteric and regulatory roles of ubiquitin [11,21]. To date, the development of a general protocol for protein ubiquitination seems still far from being in the reach but it is possible that, given the diversity of the targets and of the enzymes involved, generalization is intrinsically not possible. Our work sets an important proof-of-concept which demonstrates that it is feasible to obtain milligrams of a mono-ubiquitinated protein with a native isopeptide bond through large scale in vitro ubiquitination. The homogeneous product is suitable for a structural characterization by X-ray crystallography and/or NMR. In particular, as shown here, different 15N and 13C isotope labelling schemes can easily be introduced in the final product to perform NMR studies and address specific questions.
A side observation that could have more general applications is that the DUB activity of Josephin helps reduce the formation of long poly-ubiquitinated chains. Thus, we suggest that Josephin and/or other DUBs could potentially be exploited for in vitro ubiquitination of other substrates. Addition of even small quantities of these DUBs could help to increase the mono-ubiquitination yields and have useful biotechnological applications.
In conclusion, the methodology described here constitutes the first step towards a thorough characterization of the properties of mono-ubiquitated Josephin. Assignment of the NMR spectrum of mono-ubiquitinated Josephin may now provide further details on the mode of interaction of Josephin with ubiquitin covalently linked to K117. Crystallization trials of mono-ubiquitinated Josephin have also been initiated. This work will open entirely new avenues to unveil the mechanism underlying the activation of ataxin-3 DUB activity induced by ubiquitination.
3. Materials and methods
3.1. Protein expression
The N-terminal Josephin domain of ataxin-3 (residues 1–182) having all lysines but K117 mutated to arginines (JosK117-only) was produced as reported previously [28,29]. His-tagged human E1 expressed in insect cells was purchased from the Monoclonal Antibody/Protein Expression facility of the Baylor College of Medicine, Houston, Texas. His-tagged UbcH5a (E2) was expressed in BL21(DE3) E. coli using the Addgene plasmid 15782. CHIP (E3) in vector pGEX6P1 was produced as GST-tagged. Purification of E1 and UbcH5a was performed on a Ni-NTA resin (Qiagen). CHIP was purified using a Glutathione Sepharose matrix (GE Healthcare) and cleaved from GST with Prescission Protease (GE Healthcare). Commercial purified enzymes were purchased from Enzo Life Science (E1 and UbcH5a) and Merck Millipore (CHIP). Recombinant wild-type human ubiquitin was expressed as untagged protein and purified by anion exchange using a Q Sepharose resin followed by gel filtration on a Sephadex G-100 column (GE Healthcare). 15N labelled Josephin for NMR experiments was obtained by expression in minimal medium containing 15NH4Cl as the sole nitrogen source.
3.2. Optimization of the conditions for Josephin ubiquitination
Small scale in vitro ubiquitination (total volume 100 μl) was performed using commercial enzymes at the concentrations indicated in Todi et al. [27]. Reactions were run at 25 °C for 20 h. Josephin, ubiquitin, E1, UbcH5a and CHIP concentrations were scaled-up to achieve a larger production of mono-ubiquitinated Josephin. Final concentrations were 50 μM JosK117-only, 1 μM E1, 8 μM UbcH5a, 8 μM CHIP, 250 μM Ub. Recombinant enzymes expressed and purified in the lab were used at the same concentrations.
3.3. Large scale in vitro ubiquitination
The reaction was carried out at 25 °C for 20 h using 50 μM JosK117-only, 1 μM E1, 8 μM UbcH5a, 8 μM CHIP, 250 μM Ub, 4.5 mM ATP, 4.5 mM MgCl2, in buffer 50 mM Tris pH 7.5, 0.5 mM DTT. The total volume used typically ranged between 5 and 15 ml. JosK117-only protected with IAA was obtained after treatment with 20 mM IAA for 1 h in the dark. The sample was dialysed in 20 mM Na phosphate buffer, pH 6.5 to eliminate unreacted IAA.
3.4. Purification of mono-ubiquitinated Josephin
Mono-ubiquitinated JosK117-only was purified from the ubiquitination mixture by anion exchange using a 5 ml Hi-Trap Q HP column (GE Healthcare). The mixture was directly loaded on the column using buffer 50 mM Tris–HCl pH 8.0, 2 mM DTT. After a first wash step at 0.1 M NaCl, the protein was eluted with a linear salt gradient of 0.1–0.3 M NaCl, with a flux of 1.5 ml/min, for a total length of 180 min. The purified product run in a SDS–PAGE gel was digested with trypsin and analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) using an Orbitrap mass analyzer [28].
3.5. NMR spectroscopy
Mono-ubiquitinated Josephin (15N labelled Josephin linked to unlabelled ubiquitin) was prepared in 20 mM Na phosphate, pH 6.5, 2 mM DTT at a concentration of 300 μM. Measurements were performed at 25 °C on a Bruker 700 MHz spectrometer.
Acknowledgements
We thank Steve Howell for acquiring and analyzing MS data. The work was supported by NIH and MRC grants (SVT: R00 NS064097; HLP: R01 NS038712).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Lothrop A.P., Torres M.P., Fuchs S.M. Deciphering post-translational modification codes. FEBS Lett. 2013;587:1247–1257. doi: 10.1016/j.febslet.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldknopf I.L., Taylor C.W., Baum R.M., Yeoman L.C., Olson M.O., Prestayko A.W., Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 3.Sadowski M., Suryadinata R., Tan A.R., Roesley S.N., Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 2012;64:136–142. doi: 10.1002/iub.589. [DOI] [PubMed] [Google Scholar]
- 4.Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A., Heller H., Elias S., Haas A.L., Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA. 1980;77:1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershko A., Ciechanover A., Heller H., Haas A.L., Rose I.A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. USA. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z.J., Sun L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Mocciaro A., Rape M. Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control. J. Cell Sci. 2012;125:255–263. doi: 10.1242/jcs.091199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergink S., Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 10.Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 11.Komander D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 12.Hu M., Li P., Li M., Li W., Yao T., Wu J.W., Gu W., Cohen R.E., Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 13.Misaghi S., Galardy P.J., Meester W.J., Ovaa H., Ploegh H.L., Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 14.Wiener R., Zhang X., Wang T., Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng N., Schulman B.A., Song L., Miller J.J., Jeffrey P.D., Wang P., Chu C., Koepp D.M., Elledge S.J., Pagano M., Conaway R.C., Conaway J.W., Harper J.W., Pavletich N.P. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 16.Kamadurai H.B., Souphron J., Scott D.C., Duda D.M., Miller D.J., Stringer D., Piper R.C., Schulman B.A. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maspero E., Valentini E., Mari S., Cecatiello V., Soffientini P., Pasqualato S., Polo S. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 2013;20:696–701. doi: 10.1038/nsmb.2566. [DOI] [PubMed] [Google Scholar]
- 18.Olsen S.K., Lima C.D. Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer. Mol. Cell. 2013;49:884–896. doi: 10.1016/j.molcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hejjaoui M., Haj-Yahya M., Kumar K.S., Brik A., Lashuel H.A. Towards elucidation of the role of ubiquitination in the pathogenesis of Parkinson's disease with semisynthetic ubiquitinated alpha-synuclein. Angew. Chem. Int. Ed. Engl. 2011;50:405–409. doi: 10.1002/anie.201005546. [DOI] [PubMed] [Google Scholar]
- 20.McGinty R.K., Kim J., Chatterjee C., Roeder R.G., Muir T.W. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spasser L., Brik A. Chemistry and biology of the ubiquitin signal. Angew. Chem. Int. Ed. Engl. 2012;51:6840–6862. doi: 10.1002/anie.201200020. [DOI] [PubMed] [Google Scholar]
- 22.Virdee S., Ye Y., Nguyen D.P., Komander D., Chin J.W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 23.Virdee S., Kapadnis P.B., Elliott T., Lang K., Madrzak J., Nguyen D.P., Riechmann L., Chin J.W. Traceless and site-specific ubiquitination of recombinant proteins. J. Am. Chem. Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keren-Kaplan T., Attali I., Motamedchaboki K., Davis B.A., Tanner N., Reshef Y., Laudon E., Kolot M., Levin-Kravets O., Kleifeld O., Glickman M., Horazdovsky B.F., Wolf D.A., Prag G. Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J. 2012;31:378–390. doi: 10.1038/emboj.2011.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicastro G., Menon R.P., Masino L., Knowles P.P., McDonald N.Q., Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc. Natl. Acad. Sci. USA. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weeks S.D., Grasty K.C., Hernandez-Cuebas L., Loll P.J. Crystal structure of a Josephin-ubiquitin complex: evolutionary restraints on ataxin-3 deubiquitinating activity. J. Biol. Chem. 2011;286:4555–4565. doi: 10.1074/jbc.M110.177360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todi S.V., Winborn B.J., Scaglione K.M., Blount J.R., Travis S.M., Paulson H.L. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todi S.V., Scaglione K.M., Blount J.R., Basrur V., Conlon K.P., Pastore A., Elenitoba-Johnson K., Paulson H.L. Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117. J. Biol. Chem. 2010;285:39303–39313. doi: 10.1074/jbc.M110.181610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicastro G., Masino L., Frenkiel T.A., Kelly G., McCormick J., Menon R.P., Pastore A. Assignment of the 1H, 13C, and 15N resonances of the Josephin domain of human ataxin-3. J. Biomol. NMR. 2004;30:457–458. doi: 10.1007/s10858-004-4343-3. [DOI] [PubMed] [Google Scholar]
- 30.Masino L., Nicastro G., Menon R.P., Dal Piaz F., Calder L., Pastore A. Characterization of the structure and the amyloidogenic properties of the Josephin domain of the polyglutamine-containing protein ataxin-3. J. Mol. Biol. 2004;344:1021–1035. doi: 10.1016/j.jmb.2004.09.065. [DOI] [PubMed] [Google Scholar]
- 31.Bosanac I., Phu L., Pan B., Zilberleyb I., Maurer B., Dixit V.M., Hymowitz S.G., Kirkpatrick D.S. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J. Mol. Biol. 2011;408:420–431. doi: 10.1016/j.jmb.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Nicastro G., Todi S.V., Karaca E., Bonvin A.M., Paulson H.L., Pastore A. Understanding the role of the Josephin domain in the PolyUb binding and cleavage properties of ataxin-3. PLoS One. 2010;5:e12430. doi: 10.1371/journal.pone.0012430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plechanovova A., Jaffray E.G., Tatham M.H., Naismith J.H., Hay R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker R., Lewis S.M., Sasaki A.T., Wilkerson E.M., Locasale J.W., Cantley L.C., Kuhlman B., Dohlman H.G., Campbell S.L. Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat. Struct. Mol. Biol. 2013;20:46–52. doi: 10.1038/nsmb.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J., Ai Y., Wang J., Haracska L., Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- 36.Freudenthal B.D., Gakhar L., Ramaswamy S., Washington M.T. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat. Struct. Mol. Biol. 2010;17:479–484. doi: 10.1038/nsmb.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z., Zhang S., Lin S.H., Wang X., Wu L., Lee E.Y., Lee M.Y. Structure of monoubiquitinated PCNA: implications for DNA polymerase switching and Okazaki fragment maturation. Cell Cycle. 2012;11:2128–2136. doi: 10.4161/cc.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]


