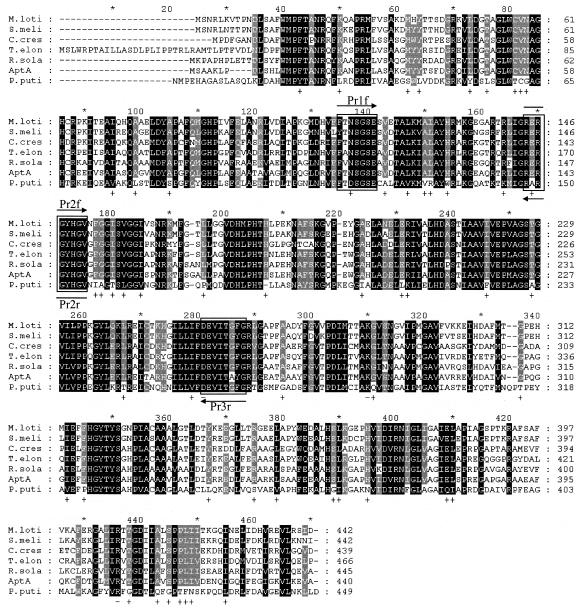
FIG. 1.
Multiple sequence alignment of A. denitrificans Y2k-2 AptA and other related proteins. Highly conserved residues are in black, and less strongly conserved residues are in gray. The boxes indicate the consensus amino acid sequences used to design the degenerate primers. M.loti, β-alanine:pyruvate transaminase from M. loti (mll1632; GenBank accession no. NP103175; 56% identity); S.meli, putative ω-APT from S. meliloti (SMc01534; GenBank accession no. NP386510; 58% identity); C.cres, ω-APT from C. crescentus (cc3143; GenBank accession no. AAK25105; 56% identity); T.elon, ω-APT from T. elongatus BP-1 (tlr0408; GenBank accession no. NP681198; 59% identity); R.sola, probable β-alanine:pyruvate transaminase from R. solanacearum (RSp1056; GenBank accession no. NP5222617; 64% identity); AptA, AptA from A. denitrificans Y2k-2; P.puti, ω-APT from P. putida (45% identity).