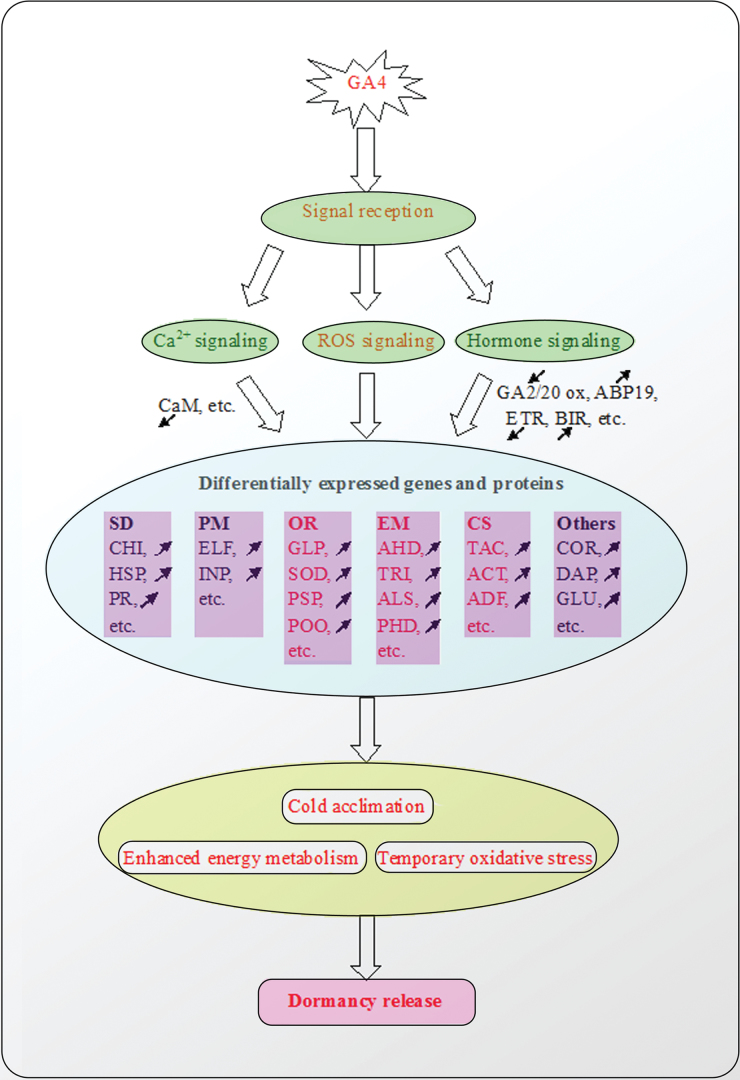
Fig. 6.
Molecular model of dormancy release in Japanese apricot treated with GA4. In this model, after GA4 treatment, signal reception, including Ca2+ signalling, ROS signalling, and hormone signalling modulate the expression of many kinds of genes and proteins, which include SD (stress and defence), PM (protein metabolism), OR (oxidation–reduction), EM (energy metabolism), CS (cell structure), and ‘others’, including signalling and transcription. These genes and proteins provide increased cold tolerance and enhanced energy metabolism to Japanese apricot, and/or cause it to enter a temporary oxidative stress state, which contributes to dormancy release. The black arrows indicate up-/downregulation of genes after GA4 treatment, and processes marked in red are more critical due to many genes or proteins being differentially expressed after GA4 treatment. Proteins and genes associated with SD, PM, OR, EM, and CS are mainly in the protein database, while those classified as ‘others’ are mainly in the DGE database. CHI, class IV chitinase; HSP, heat shock protein 60; PR, pathogenesis-related thaumatin superfamily protein; ELF, elongation factor Tu (ISS); INP, insulinase (peptidase family M16) protein; GLP, glutathione peroxidase 6; SOD, manganese superoxide dismutase and copper/zinc-superoxide dismutase; PSP, peroxidase superfamily protein; POO, polyphenol oxidase; AHD, alcohol dehydrogenase 1; TRI, triosephosphate isomerase, putative; ALS, aldolase superfamily protein; PHD, D-3-phosphoglycerate dehydrogenase; TAC, tubulin α-2 chain; ACT, actin 7; ADF, actin-depolymerizing factor; DAP, dormancy-associated protein-like 1; GLU, β-1,3-glucanase 1; ABP19, auxin-binding protein ABP19b; ETR, ethylene receptor; BIR, brassnosteroid insensitive 1- associated receptor kinase 1 precursor, putative. (This figure is available in colour at JXB online.)