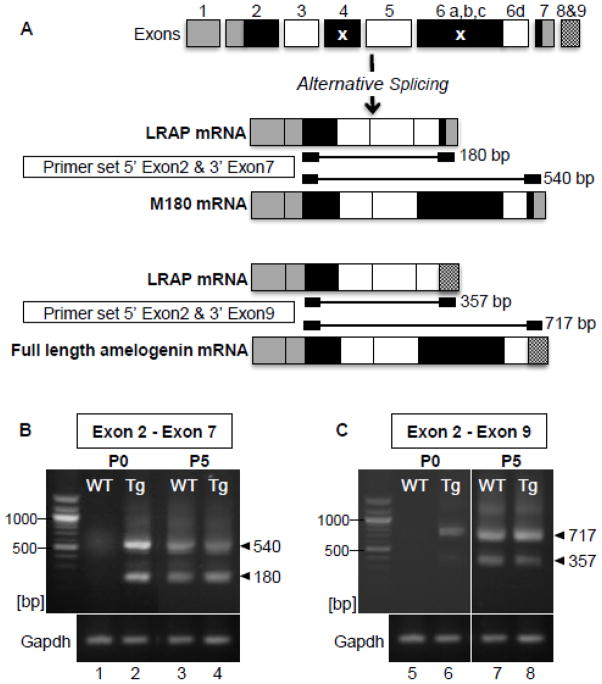
Figure 4. Expression of multiple spliced variants of amelogenin was upregulated in molars from TgLRAP mice at P0.
(A) Spliced variants of amelogenin mRNA and PCR products size using two different primer sets are shown. LRAP lacks exon 4 and the majority of exon 6 (6 a, b, and c) as indicated by x. Inclusion of exons 8 and 9, occurs when exon 7 is sliced out, ending with the alternative stop codons in exon 9 (LI et al). (B) In P0 WT mice, M180 mRNA expression was barely detected (lane 1), whereas in P0 TgLRAP mice both LRAP (180 bp) and M180 (540 bp) variants were clearly upregulated compared to WT (lane 2). At P5, both splice variants were observed in WT and TgLRAP with a similar relative intensity (lane 3 and 4). Sequencing results confirmed the dominant upper band from lane 2 matched the M180 amelogenin. (C) A primer set (5′ exon 2 and 3′ exon9) allowed detection of endogenous amelogenin mRNA but not the LRAP transgene. At P0, RT-PCR products for full length amelogenin (717 bp) and LRAP8,9 (357 bp) were observed only in TgLRAP mice(lane 6) but not WT (lane 5). At P5, both WT and TgLRAP mice showed two spliced variants at similar level (lane 7 and 8).