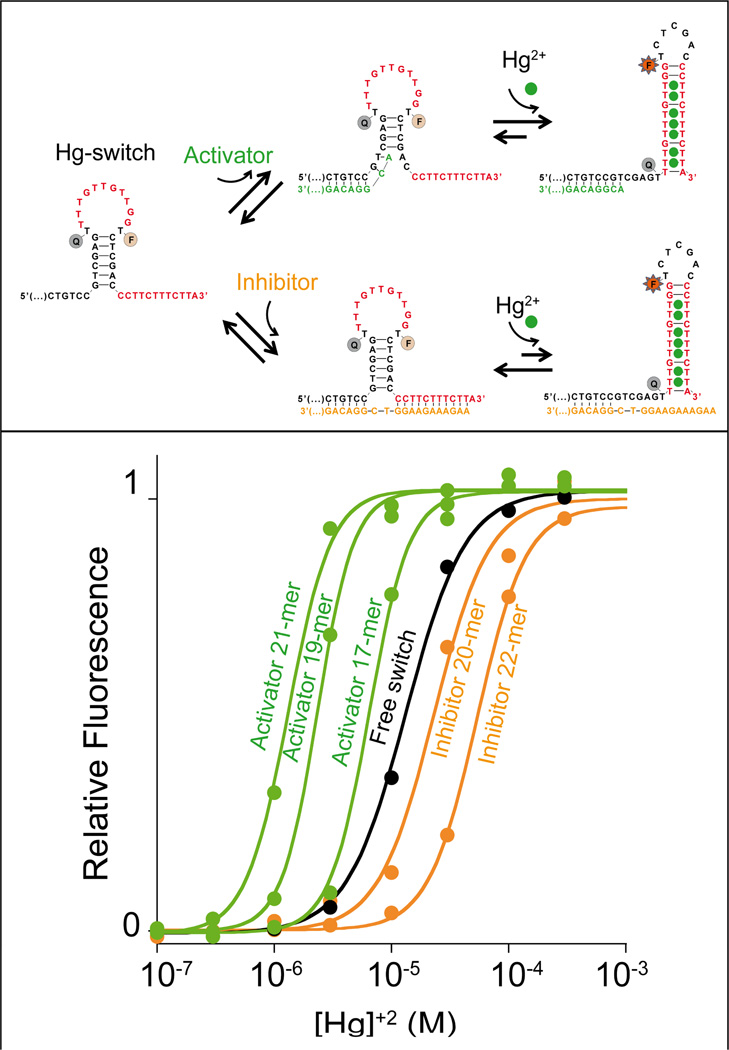
Figure 3.
We can tune the dynamic range of metal-activated DNA-switches using heterotropic allostery in which the addition of allosteric effectors (activator/inhibitor) pushes the useful dynamic range to higher or lower target concentrations. Top: As an allosteric activator we employ a single-stranded DNA that, when bound to a tail appended to the 5’ end of the switch, destabilizes the non-binding conformation and thus pushes the useful dynamic range of the switch to lower target concentrations (bottom, green curves). As an allosteric inhibitor, in contrast, we employed a sequence that binds the tails on both ends of the switch simultaneously, thus stabilizing the non-binding conformation and pushing the dynamic range of the switch to higher target concentrations (bottom, orange curves). Bottom: Increasing the length of the inhibitor from 20 to 22 bases increasingly stabilizes the non-binding state thus pushing the useful dynamic range to higher and higher target concentrations (orange curves). Increasing the length of the activator from 17 to 21 bases similarly increases the extent to which it destabilizes the non-binding state, pushing the useful range of the switch to lower target concentrations (green curves). The black curve (free switch) represents the binding of the switch in the absence of allosteric effectors. Binding curves are shown as normalized signals. We note, however, that, because activator binding destabilizes the non-binding conformation we observe an increase of the background signal. As a result, in the presence of activators, we obtain a lower signal gain in response to the target (Figure S4). We also note that no significant quenching of mercury(II) ions on the FAM signal was observed (Figure S5). Here the switch concentration was held at 10 nM and the activator or inhibitor concentration at 20 nM, a concentration that allows saturation of the switch (see figure S6 and SI for more details).