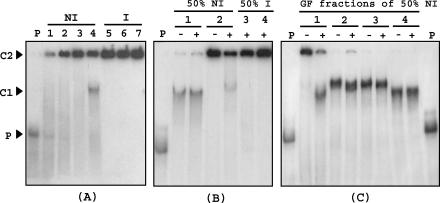
FIG. 6.
Mobility shift assay of a DNA probe corresponding to the padA promoter region (2 × 10−8 M), generated and labeled by PCR with [α-32P]dATP, with crude protein extracts, partially purified with (NH4)2SO4 and by gel filtration, from noninduced (NI) or p-coumaric acid-induced (I) L. plantarum cells. (A) Binding assay with (NH4)2SO4 fractions from NI or I cells. P, DNA probe without protein extract; lane 1, crude extract from NI cells; lanes 2 and 5, 30% (NH4)2SO4 fraction; lanes 3 and 6, 40% (NH4)2SO4 fraction; lanes 4 and 7, 50% (NH4)2SO4 fraction. (B) Binding assays with 50% (NH4)2SO4 fractions from NI and I cells preincubated with (+) or without (−) 3 × 10−7 M unlabeled ldhL promoter DNA as a competitor (see Materials and Methods). P, DNA probe without protein extract; lanes 1 and 3, 0.025 μg of protein/μl; lanes 2 and 4, 0.075 μg of protein/μl. (C) Binding assays with the four pools of protein obtained by gel filtration (GF) of the 50% (NH4)2SO4 protein fraction from the NI protein extract with (+) or without (−) ldhL promoter DNA as a competitor. P, probe without protein extract; lanes 1, 2, 3, and 4, respectively, the first, second, third, and fourth pools of proteins collected by gel filtration elution. The protein concentration of the pools was the same as that in the binding assays (0.025 μg/μl). C1, band corresponding to a specific binding of the probe with a protein in extracts or protein fractions exclusively from NI cells; C2, band corresponding to the binding of the probe with a high-molecular-mass protein.