Figure 1.
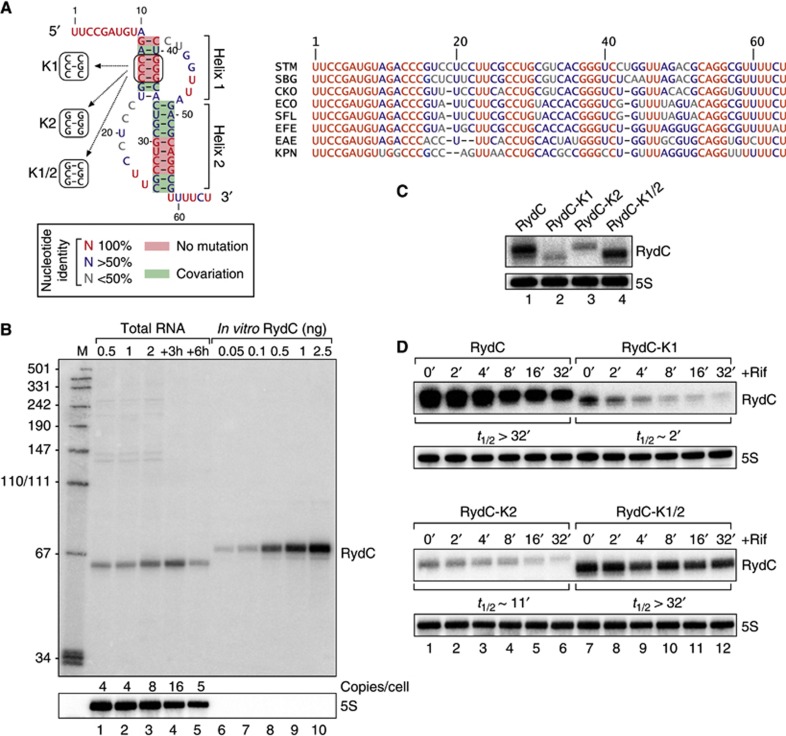
Conservation, structure and expression of RydC. (A) Alignment and pseudoknot structure of RydC RNA. STM: Salmonella Typhimurium; SBG: Salmonella bongori; CKO: Citrobacter koseri; ECO: Escherichia coli; SFL: Shigella flexneri; EFE: Escherichia fergusoni; EAE: Enterobacter aerogenes; KPN: Klebsiella pneumoniae. Mutations K1, K2 and K1/2 introduced to alter pseudoknot formation are indicated. (B) RydC levels in total RNA samples (corresponding to 1 OD600) of wild-type Salmonella at indicated time points of growth were compared on northern blots to signals of in vitro transcribed RydC to estimate the in vivo copy number. 5S RNA served as a loading control. (C) Expression levels of RydC, RydC-K1, RydC-K2 and RydC-K1/2 (as described in (A); expressed from the constitutive PL promoter) were determined in ΔrydC Salmonella by northern blot analysis of total RNA samples (OD600 of 1). (D) Stabilities of RydC, RydC-K1, RydC-K2 and RydC-K1/2 were determined in ΔrydC Salmonella by northern blot analysis of total RNA samples withdrawn prior to and at indicated time points after inhibition of transcription by rifampicin at an OD600 of 1. See Supplementary Figure S4 for quantification.
Source data for this figure is available on the online supplementary information page.