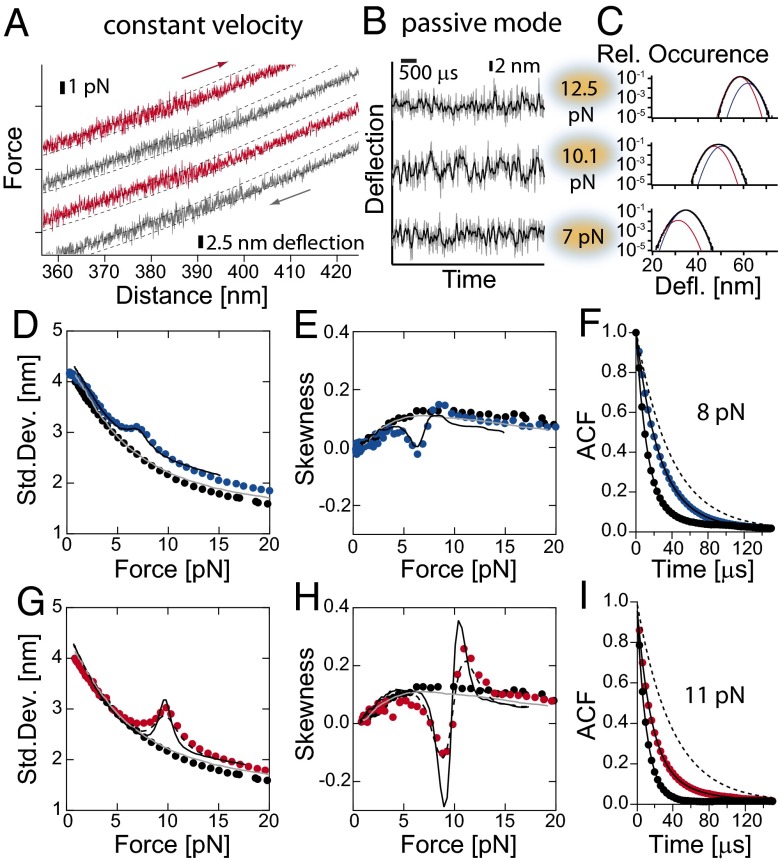
Fig. 2.
(A) Force-distance traces for HP35stab at 30 kHz. Red traces are stretch cycles and the gray traces are relax cycles. For kinetic analysis, the positions of both traps were fixed and set at different forces. (B) Time trace of deflection fluctuations of HP35stab at 12.5, 10.1, and 7 pN as measured at 300 kHz (gray lines) and at 30 kHz (black lines), and (C) corresponding histograms of deflections. Histograms were fitted by assuming two skewed Gaussian distributions: for folded state (blue line) and for unfolded state (red line) and the sum of the both Gaussian curves (black line). From the distribution, we extracted describing parameters such as SD and skewness. (D, E, G, and H) Continuous lines are results from Brownian dynamics simulations (SI Appendix). The solid gray line is obtained from a simulation describing the bead-DNA-bead system assuming linker parameters as obtained from experiments. The black solid line is the result of a simulation of the bead-DNA-protein-DNA-bead system assuming linker parameters as obtained from experiments. The protein folding/unfolding transitions were modeled as a two-state system or a three-state system in the case of HP35stab (black dashed line) with Markov chain properties (Table 1 and SI Appendix, Figs. S5 and S4). (D and G) SD of fluctuations in bead deflection as function of mean force. Shown are DNA construct (black circles), HP35WT (blue circles), and HP35stab (red circles). (E and H) Skewness of fluctuations in bead deflection as function of mean force. Shown are data for a DNA construct (black circles), HP35WT (blue circles) and HP35stab (red circles). (F) Autocorrelation function of the deflection signal of DNA (black circles) and HP35WT at 8.0 pN (red circles). For comparison, we also show the autocorrelation function for untethered beads (dashed line). The solid lines are curve fits to a single- or double-exponential decay function. Analysis gave time constant for untethered beads of τ0 = 56 µs, for a DNA-only tether τ0 =13 µs and for an HP35WT tether τ0 = 13 µs and τ1 = 32 µs, respectively. (I) Autocorrelation function of the deflection signal of DNA (black circles) and HP35stab at 11.0 pN (red circles). For comparison, the autocorrelation function for untethered beads is also shown (dashed line). The solid lines are curve fits of a single- or double-exponential equation to the data. Analysis resulted in a time constant of τ0 = 56 µs for untethered beads, for DNA τ0 = 11 µs, and for HP35stab τ0 = 14 µs and τ1 = 30 µs with τ0 as a free fitting parameter.