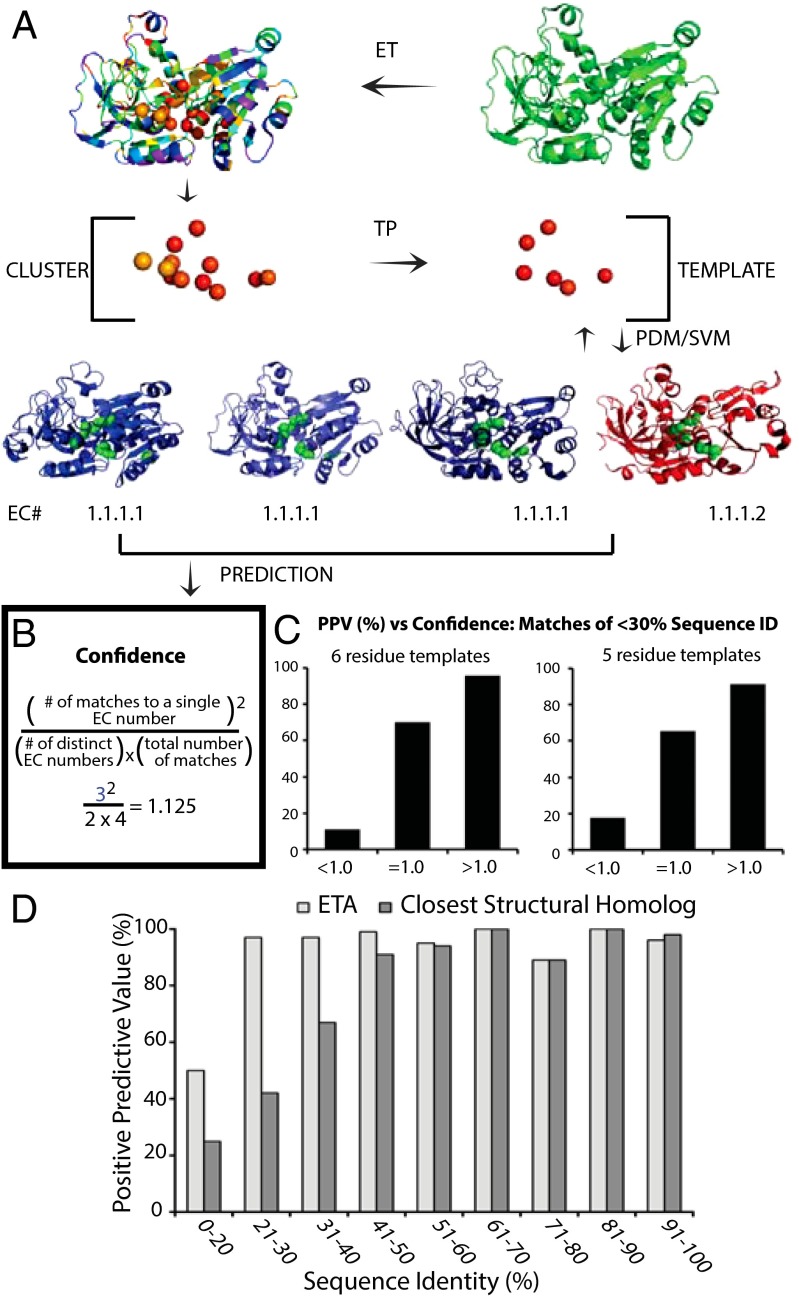
Fig. 1.
ETA accurately determines substrate specificity. (A) The ET algorithm is applied to a protein from Sulfolobus tokadaii strain 7 (green, PDB ID code 2eer, chain A) to identify evolutionarily important residues. A cluster of 10 or more important residues is identified and a Template Picker algorithm further selects five or six residues to act as a template that is used to probe a target library of proteins with known functions. Paired-distance matching algorithm identifies regions in protein structures in the target library that are similar to the template. Found matches are next passed to the SVM, which identifies significant matches based on geometric and evolutionary similarities. ETA repeats all these steps reciprocally, generating templates from target structures and searching for matches in the query protein. Following this protocol, ETA suggests four matches: alcohol dehydrogenase from Saccharomyces cerevisae (blue left, PDB ID code 2hcy), alcohol dehydrogenase from S. solfataricus (blue middle, PDB ID code 1r37), human class II alcohol dehydrogenase (blue right, PDB ID code 3cos), and NADP(H)-dependent cinnamyl alcohol dehydrogenase from S. cerevisae (red, PDB ID code 1piw) to the query protein. (B) The most seen function among matches, alcohol dehydrogenase activity (EC 1.1.1.1), is identified with high confidence with a confidence value of 1.125 as calculated in the box. (C) Comparison of PPV versus confidence score binned at <1, =1, and >1 for both six-residue templates (Left) and five-residue templates (Right) when considering only matches of <30% sequence identity. For more detail, see Fig. S1. (D) Comparison of PPV when predictions are made using ETA or the closest structural match (TM-align). Horizontal axis shows the maximum sequence identity of matches for proteins depicted in corresponding bars; the vertical axis is the PPV for each bin range.