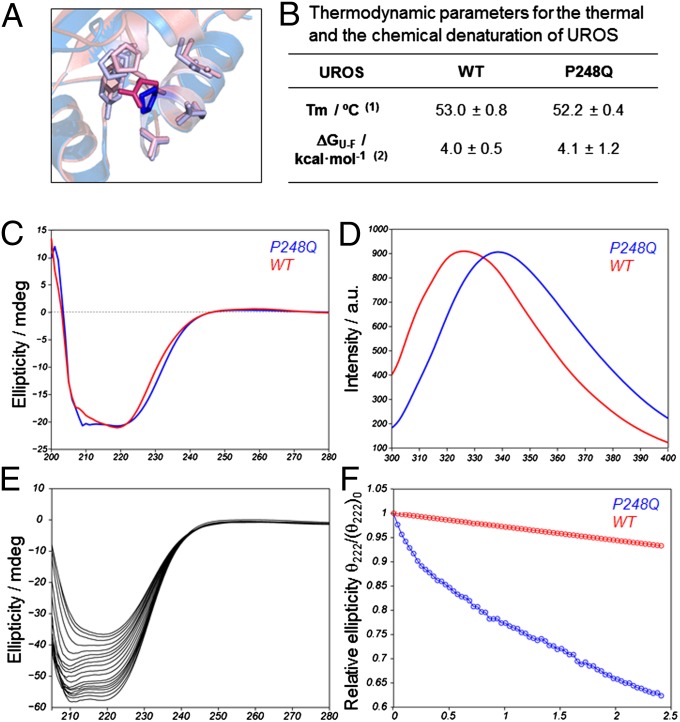
Fig. 1.
Reduced kinetic stability of UROSP248Q in vitro. (A) Overlay of the WT UROS X-ray structure (1jr2, blue) and the modeled structure for UROSP248Q (pink). The highlighted side chain corresponds to the mutation site. This side chain forms contacts with other nearby residues, also highlighted: P18, Y19, E22, P246, T247, Q249, L251, and A252. (B) Thermodynamic parameters for the thermal and the chemical denaturation of UROSP248Q. (1) Melting temperature (Tm) monitoring the secondary structure change by circular dichroism at 1 °C/min. (2) Unfolding free energy in the absence of denaturant. (C and D) Far utraviolet-circular dichroicism spectrum (C) and natural abundance tryptophan emission spectrum (D) for the two proteins under consideration. (E) Evolution of the folded conformation over time at physiological temperature. The spectral change reflects the loss of α-helical (signal at 222 nm) toward β-sheet (signal at 210 nm) conformation over time for UROSP248Q. (F) Signal decay over time (at 222 nm) for UROSWT (red) and UROSP248Q (blue).