Abstract
Anion and cation effects on the structural stability of lysozyme were investigated using differential scanning calorimetry. At low concentrations (<5 mM) anions and cations alter the stability of lysozyme but they do not follow the Hofmeister (or inverse Hofmeister) series. At higher concentrations protein stabilization follows the well-established Hofmeister series. Our hypothesis is that there are three mechanisms at work. At low concentrations the anions interact with charged side chains where the presence of the ion can alter the structural stability of the protein. At higher concentrations the low charge density anions perchlorate and iodide interact weakly with the protein. Their presence however reduces the Gibbs free energy required to hydrate the core of the protein that is exposed during unfolding therefore destabilizing the structure. At higher concentrations the high charge density anions phosphate and sulfate compete for water with the protein as it unfolds increasing the Gibbs free energy required to hydrate the newly exposed core of the protein therefore stabilizing the structure.
Keywords: calorimeter, DSC, formulation, Hofmeister, chaotrope, kosmotrope
Introduction
In 1888 Franz Hofmeister discovered that different salts had different effects on the solubility of protein ensembles found in hen egg white.1 This effect was found to be consistent for a number of protein ensembles and a ranking order showing the effect of different anions and cations on protein solubility was developed. Ions that cause salting-out (precipitation) were termed kosmotropes (water structure makers).2 Ions that cause salting-in were termed chaotropes (water structure-breakers). An extended series for anions and cations and their effect on proteins is shown below.
Anions to the left of chloride have a higher charge density and decrease protein solubility.3–5 Anions to the right of chloride have a lower charge density and increase protein solubility. Some of the earliest pioneering work on how the salts of the Hofmeister series affect protein stability was carried out by Peter von Hippel and Kwok-Ying Wong in 1965.6 In this work the effect of high salt concentrations (1–4 M) on the thermal stability of ribonuclease A was measured for a variety of different salts. The experiments showed that the effect of salts on protein stability followed the same order as the Hofmeister series. High charge density ions stabilized and the low charge density ions destabilized the protein. Originally it was thought that the kosmotropic and chaotropic effects were due to the ion's ability to make or break water structure around proteins.4 Femtosecond pump-probe spectroscopy and pressure perturbation calorimetry have generated evidence suggesting that the theory of ions making or breaking water structure is not necessarily true.7–9 Femtosecond pump-probe spectroscopy showed that anions form hydration shells around themselves irrespective of charge density and that the hydration shell does not extend past one water layer for either. It also showed that high charge density ions bind their layer of water more tightly than low charge density ions. Preferential interaction between salts and proteins displacing water from the interface has been linked to protein stability.10 Despite the effects of the Hofmeister series being observed for 125 years a definitive molecular-based mechanism is not known.
There is a body of work which studies the role salts play during the unfolding of proteins induced by urea,11–13 guanidinium HCl,14 low pH,15,16 and heat.17–22 The role of salts during heat-induced unfolding is not clear from the literature. Research on the effect of the anions on unfolding of the B1 domain of protein L demonstrated that sulfate, phosphate and fluoride stabilized, while chloride was neutral and nitrate, perchlorate and thiocyanide destabilized the protein.18 The stabilization was measured as the change in the temperature of maximum unfolding (ΔTm). The ΔTm was proportional to the cosolute concentration and agreed with the findings of von Hippel and Schleich. NMR experimentation also showed a significant shift of the amides in the protein in the presence of the low charge density anion thiocyanide. This was explained in terms of stabilization by excluded volume which hampers unfolding and an opposite preferential anion solvation (weak binding of the cosolute to the protein surface) that destabilizes the protein. Thermal unfolding of ribonuclease A differed from protein L in that low concentrations of sulfate (around 1 mM) rapidly stabilized the protein followed by a slower rise in stability17 suggesting that two phases of stabilization occur. The two phases suggest a degree of complexity to protein stabilization by anions that cannot be explained by a single theory such as preferential interaction. The mechanisms for protein unfolding by urea, guanidinium HCl and low pH are sufficiently different to thermal unfolding to make extrapolation from these unfolding conditions to thermal unfolding problematic.
Cloud point analysis used to study the role of anions on lysozyme solubility identified an inverse Hofmeister effect at low ion concentrations which was linked to the size of the anion23 and its capacity to screen the charged residues on the protein. The aim of the research presented in this article was to investigate whether lysozyme stability at low anion concentrations complied with the inverse Hofmeister series and investigate the transition where the direct Hofmeister behavior starts to dominate lysozyme stability. In this article differential scanning calorimetry (DSC) was used to investigate how two high charge density anions: phosphate (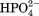 ) and sulfate (
) and sulfate (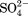 ), an intermediate anion chloride (Cl−) and two low charge density anions: iodide (I−) and perchlorate (
), an intermediate anion chloride (Cl−) and two low charge density anions: iodide (I−) and perchlorate (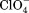 ) affect the thermal stability of lysozyme at pH 7, while maintaining sodium as the cation for all anions. Sodium was chosen because it is regarded as a neutral cation, so should have little effect on the stability of lysozyme. Keeping the cation component constant throughout the anion experiments enabled us to directly compare the effect of the different anions, without interference from other ions in solution. The anions were chosen based on their positions in the Hofmeister series. Their effects on protein stability were investigated over a 0.125 mM–1 M concentration range. Lysozyme was adjusted to pH 7.0 so it would have a net positive charge making it attractive to the negative anions. The cations caesium (Cs+), ammonium (
) affect the thermal stability of lysozyme at pH 7, while maintaining sodium as the cation for all anions. Sodium was chosen because it is regarded as a neutral cation, so should have little effect on the stability of lysozyme. Keeping the cation component constant throughout the anion experiments enabled us to directly compare the effect of the different anions, without interference from other ions in solution. The anions were chosen based on their positions in the Hofmeister series. Their effects on protein stability were investigated over a 0.125 mM–1 M concentration range. Lysozyme was adjusted to pH 7.0 so it would have a net positive charge making it attractive to the negative anions. The cations caesium (Cs+), ammonium (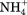 ), sodium (Na+), potassium (K+), magnesium (Mg2+), and calcium (Ca2+) were then tested using chloride as the anion. The lysozyme was dialyzed against pure water prior to DSC experiments to remove counter ions present in the purchased material. No buffer was used so that removal of charged buffer components from the protein surface did not interfere with the analysis. The natural buffering capacity of the protein was used to stabilize the pH of the preparation. In some previous studies where buffers were used it is not clear whether the addition of the ion or removal of the buffering ion was being studied.
), sodium (Na+), potassium (K+), magnesium (Mg2+), and calcium (Ca2+) were then tested using chloride as the anion. The lysozyme was dialyzed against pure water prior to DSC experiments to remove counter ions present in the purchased material. No buffer was used so that removal of charged buffer components from the protein surface did not interfere with the analysis. The natural buffering capacity of the protein was used to stabilize the pH of the preparation. In some previous studies where buffers were used it is not clear whether the addition of the ion or removal of the buffering ion was being studied.
Results and Discussion
Thermal analysis was undertaken using a capillary DSC where convection is minimal and sensitivity is high enough to detect unfolding at low protein concentrations. The peak in the DSC scan is referred to as the temperature of maximum unfolding (Tm) and was used as the measure of the thermal stability of lysozyme in this experiment (Fig. 1). The higher the Tm value the more stable the protein structure. The precision of the DSC was high (standard deviation of 0.02°C) and the reproducibility was also good as demonstrated by the repeated NaCl experiment [Figs. 2(a) and 4].
Figure 1.
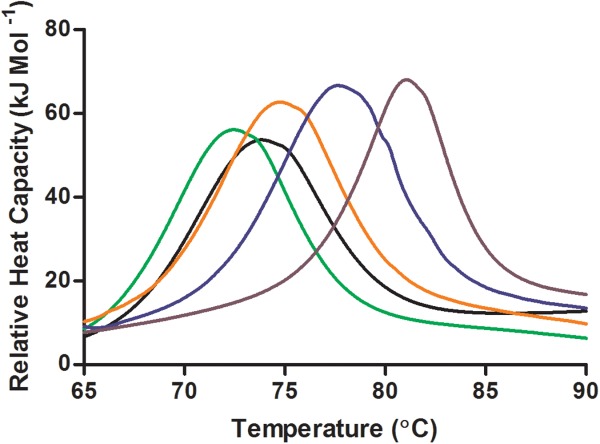
Differential scanning calorimetry scans of lysozyme with varying sodium phosphate concentrations, 0 mM (black), 1 mM (green), 250 mM (orange), 500 mM (blue), and 1000 mM (purple), all at pH 7.0.
Figure 2.
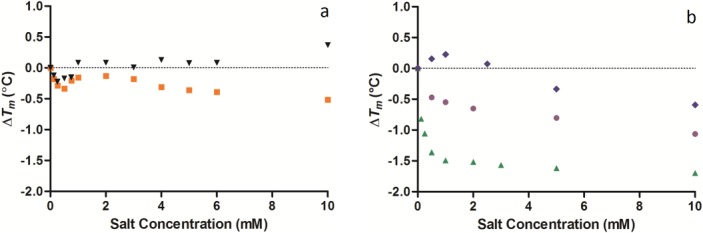
Change in the temperature of maximum unfolding (ΔTm) values of lysozyme measured by DSC with varying concentrations of the anions (a) chloride ( ), iodide (
), iodide ( ), (b) sulfate (
), (b) sulfate ( ), phosphate (
), phosphate ( ), and perchlorate (
), and perchlorate ( ) between 0 and 10 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
) between 0 and 10 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 4.
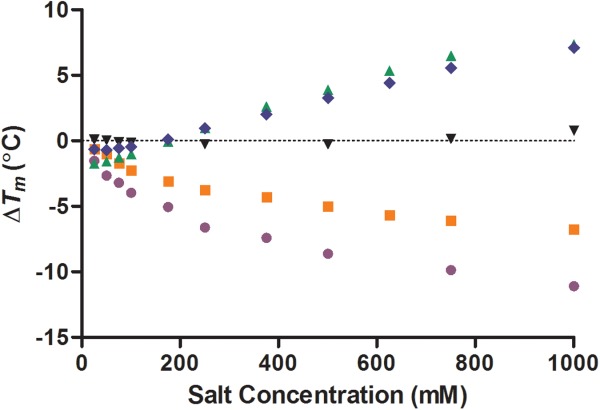
Change in the temperature of maximum unfolding (ΔTm) values of lysozyme measured by DSC with varying concentrations of the anions chloride ( ), iodide (
), iodide ( ), sulfate (
), sulfate ( ), phosphate (
), phosphate ( ), and perchlorate (
), and perchlorate ( ) between 10 and 1000 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
) between 10 and 1000 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Lysozyme thermal stability at low salt concentrations (0–10 mM)
Chloride is regarded as an intermediate anion in the Hofmeister series. It had an effect on the stability of lysozyme at low concentrations below 1 mM [Fig. 2(a)]. There is an initial drop of 0.22°C in the Tm of lysozyme with the addition of 0.25 mM NaCl. The Tm then increased 0.31°C with the addition of a further 0.75 mM NaCl to bring the total NaCl concentration to 1 mM. From 1 to 5 mM NaCl very little change in the Tm of lysozyme was observed. Iodide is a low charge density anion but had a very similar effect on lysozyme stability at low concentrations to chloride [Fig. 2(a)]. Iodide destabilized lysozyme by 0.28°C with the addition of 0.25 mM NaI and then stabilized lysozyme by 0.13°C with the addition of further 0.75 mM NaI to bring the total NaI concentration to 1 mM. After the NaI concentration exceeded 1 mM there was a slow decline in the Tm of lysozyme dropping 0.20°C between 1 and 5 mM NaI. Phosphate is a high charge density anion and behaved quite differently to chloride and iodide [Fig. 2(b)]. The Tm of lysozyme decreased by 1.49°C with the addition of 1 mM phosphate. Further addition of phosphate up to 5 mM caused very little change in the Tm of lysozyme (only 0.13°C). Sulfate is also a high charge density anion but initially did the opposite to phosphate, lysozyme was stabilized by 0.22°C with the addition of 1 mM sulfate [Fig. 2(b)], further addition of sulfate up to 5 mM caused destabilization of lysozyme by 0.56°C. Perchlorate is a low charge density anion and causes an initial drop in the Tm of lysozyme of 0.55°C with the addition of 1 mM NaClO4 [Fig. 2(b)], further addition of perchlorate to 5 mM further reduced the Tm of lysozyme by another 0.36°C. The first observation is that the stabilization of lysozyme by anions at concentrations below 1 mM is not related to the position of the anion in the Hofmeister series.
During the testing of different cations the anion chloride was used. At a concentration of 1 mM, sodium chloride [Fig. 3(a)] demonstrated the small dip in stability shown previously [Fig. 2(a)]. The other cations also presented a dip in stability [Fig. 3(a,b)] suggesting the slight change in stability is dictated by the chloride anion. The shape of the slight dip in Tm was modulated between competition between the protein and the various cations for the chloride.
Figure 3.
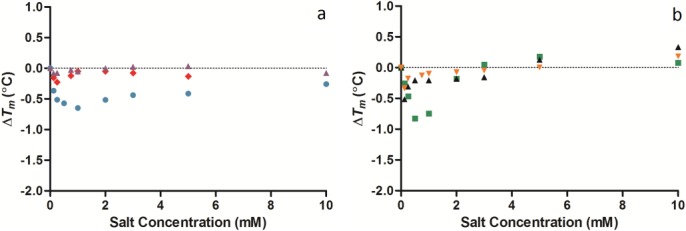
Change in the temperature of maximum unfolding (ΔTm) values of lysozyme measured by DSC with varying concentrations of the cations (a) sodium ( ), potassium (
), potassium ( ), caesium (
), caesium ( ) (b) ammonium (
) (b) ammonium ( ), calcium (▴), and magnesium (
), calcium (▴), and magnesium ( ) between 0 and 10 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
) between 0 and 10 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The effect that the five anions and six cations had on the structural stability of lysozyme at the intermediate concentration range (2–10 mM) was found to be small [Fig. 2(a,b) and 3(a,b)]. Chloride continued to have very little effect on the Tm of lysozyme, increasing the Tm by only 0.28°C with the increase in chloride from 2 to 10 mM. Phosphate also caused very little effect on lysozyme structural stability reducing the Tm by only 0.18°C. Iodide destabilized lysozyme reducing the Tm by 0.38°C. Sulfate had a similar effect to iodide, the Tm of lysozyme decrease by 0.66°C. The effect perchlorate had over 2.0–10 mM was consistent with it being a chaotrope as it caused a slow steady decrease of 0.18°C in the Tm. At these intermediate concentrations there was also no distinguishable agreement with the Hofmeister series apart from iodide and perchlorate, where the Tm of lysozyme was decreased. Sulfate, despite being a high charge density anion also destabilized lysozyme like a chaotrope. All cations affected the Tm by <0.5°C [Fig. 3(a,b)].
Lysozyme thermal stability at higher salt concentrations (10–1000 mM)
As the anion concentration was raised from 50 to 1000 mM, the high charge density anions sulfate and phosphate started to stabilize lysozyme raising the Tm by 7.80 and 8.93°C, respectively (Fig. 4). The low charge density anions perchlorate and iodide decreased lysozyme stability as the concentration was raised from 10 to 1000 mM lowering the Tm by 10.03 and 6.26°C, respectively. Chloride's stabilization of lysozyme was negligible raising the concentration from 50 to 1000 mM increased the Tm by just 0.75°C. It is over 50 mM concentration that the anions are in agreement with the Hofmeister series and the experimental findings of von Hippel and Schleich.6 The monovalent anions caesium, sodium, potassium, and ammonium had minimal effect on lysozyme stability (effecting the Tm by <1.0°C) but the divalent cation calcium destabilized lysozyme (1000 mM calcium reduced the Tm by 3.63°C) [Fig. 5(a,b)].
Figure 5.
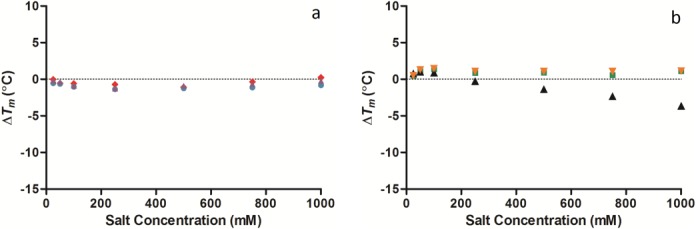
Change in the temperature of maximum unfolding (ΔTm) values of lysozyme measured by DSC with varying concentrations of the cations (a) sodium ( ), potassium (
), potassium ( ), caesium (
), caesium ( ) (b) ammonium (
) (b) ammonium ( ), calcium (▴), and magnesium (
), calcium (▴), and magnesium ( ) between 10 and 1000 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
) between 10 and 1000 mM salt concentration (all at pH 7.0). The standard deviation for the Tm of lysozyme was 0.02°C (n = 5). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Mechanisms for anion stabilization and destabilization of lysozyme
To understand the role of anions on the thermal stability of proteins the unfolding reaction should be considered. Firstly to unfold the noncovalent bonds between adjacent polypeptide chains (which include a combination of hydrogen bonds, van der Waals forces and salt bridges) need to be broken. Then the newly exposed hydrophobic core of the protein has to be hydrated. Finally, the sections with secondary structure, alpha helices and beta pleated sheets which are held together by hydrogen bonds needs to be denatured. The hydration of the newly exposed core is observed as a rise in heat capacity in DSC scans due to the heat capacity of water associated with apolar side chains being higher than the bulk water.24,25 For an ion to alter the stability of a protein it must either directly alter the strength of the protein structure, or alter the change in free energy associated with hydration of the newly exposed core. It is worth noting that thermal unfolding reactions are the reverse to folding reactions. During protein folding parts of the protein's hydration layer are displaced as the residues become buried. This is accompanied by the formation of intramolecular hydrogen bonds, van der Waal's bonds and salt bridges. Precipitation reactions also involve the displacement of part of the protein's hydration layer and the formation of intermolecular non-covalent bonds. The role of water in both protein folding and precipitation reactions has been demonstrated by their negative activation enthalpies as the water in the hydration layer is displaced.26,27 The relationship between solubility (the precipitation reaction) and stability with regards to the effect of salts has been accepted since the work of Peter von Hippel and Kwok-Ying Wong,6 which would indicate that understanding the mechanisms dictating solubility will enhance our understanding of stability (and vice versa).
Our understanding of the hydration layer around proteins (including the layer that forms around exposed core of a protein during unfolding) is still an area of active research and debate, and has been heavily influenced by the analytical approach used. Neutron scattering observes tightly bound water associated with charged or polar side chains supporting the theory of the hydration layer being a single or double layer. The understanding of the water interaction with apolar residues is more contentious. 2H NMR relaxation work suggests that water interacting with an apolar solute or interface is perturbed, being the reverse of the classical “iceberg of structured water” theory proposed by Frank and Evans in 1945.28,29 Recent studies using THz spectroscopy also indicate that the water associated with apolar side chains is more extensive than previously thought.30–32 Calorimetry also identifies a population of water associated with apolar side chains that has a relatively high heat capacity (ΔCp) and a negative relationship with temperature (−ve δCp/δT).24 As a protein unfolds the apolar interior of the protein becomes exposed to water and hydrated which increases the heat capacity.25 Irrespective of the exact nature of the hydration layer around proteins it would be safe to suggest that water around apolar residues is significantly different to that around polar and charged portions of the protein's surface. Modulating the free energy associated with hydration of the newly exposed apolar surface area by ions may be a mechanism for anion stabilization or destabilization of proteins.
At pH 7, lysozyme has a positive net charge of around +8.33 The anions may interact electrostatically with the positively charged amino acid side chains of lysine, arginine and histidine if they are accessible and not sterically hindered. This is consistent with the observed change in stability with addition of salts below 2 mM. The effect of the bound anion on the structural stability of the protein may vary depending on local conditions of the protein surface. Monovalent anions will neutralize exposed positively charged side chains, which will affect neighboring residues, ending repulsion between positively charged residues and attraction of negatively charged residues. This may result in a force being applied to the peptide backbone, stabilizing or destabilizing the protein in the process. Binding of the divalent anions to positively charged side chains will reverse the charge at this location repelling negatively charged residues and attracting positively charged residues, and similarly affect the peptide backbone. It is difficult to predict whether the presence of a bound anion will stabilize or destabilize a protein and in the case of chloride and iodide their presence did both, which was probably dependent on which side chain received the anion. The destabilization of lysozyme by phosphate mirrors that of sodium phytate (an inositol ring with six phosphate groups attached) where binding was demonstrated to be electrostatic and reversible by the addition of chloride ions.34 A study of the effect of anions on the solubility of lysozyme using cloud-point testing also suggested electrostatic interaction at lower salt concentrations that could be modelled using a Langmuir-type isotherm. Interestingly the effect on solubility followed an inverse Hofmeister series23 suggesting the mechanism by which bound anions effect stability and solubility are unrelated. Lysozyme solubility was dictated by the anion screening of the electrostatic repulsion between charged molecules. Lysozyme stability, however, cannot be explained by Debye screening alone. There is no relationship between ionic strength and stability (see Fig. 1 in Supporting Information). We propose local electrostatic forces may play a role in modulating structural stability.
The cations were studied keeping the chloride anion constant. Below 2 mM salt the effect on stability is probably dominated by the chloride anion and the cation modulated its effect. Cations with high charge density (e.g., sodium and potassium) preferentially interact with the chloride at the expense of binding to the positively charged side chains on the protein. Cations with lower charge density (e.g., ammonium and caesium) don't preferentially interact with the chloride and therefore the chloride is free to interact with the protein and alter its stability. Above 25 mM, calcium was the only cation that had significant impact on lysozyme stability. Interestingly the other divalent cation magnesium had little effect suggesting the effect of calcium on lysozyme is unique to this cation.
Over 2 mM salt concentration the low charge density anions behave in a predictable manner reducing the Tm value of the protein. It has been suggested that they can interact weakly with apolar surfaces on the protein.35 1H-15N two-dimensional heteronuclear single-quantum correlation (HQSC) spectra generated for protein L in the presence of the low charge density ion SCN− suggested a weak binding of the low charge density ion to the protein.18 We propose that during unfolding the presence of an anion that interacted with newly exposed apolar surfaces reducing the free energy required to hydrate this surface. This would reduce the total free energy of unfolding and therefore promote destabilization of the protein structure.
At around 25–50 mM salt concentration the high charge density anions start to behave in a predictable manner increasing the Tm value of the protein. High charge density anions may compete for water required to hydrate the newly exposed apolar surface area during unfolding raising the free energy for this process. 1H-15N HQSC spectra of the high charge density anions sulfate and phosphate interacting with protein L indicated no direct interaction between the anion and the protein. Molecular crowding is an accepted phenomenon where the presence of organic molecules (or macromolecules) causes a marked rise in the Tm value of a protein.36 The authors propose that it is the competition for water between the hydration layers of anions (or organic molecular crowding agents) and the protein as it unfolds that increases the Gibbs free energy of the unfolding process.
The model proposed by the authors has three mechanisms by which anions modulate lysozyme stability (at pH 7). These are: electrostatic interaction at low concentrations, weak interaction with apolar residues and competition for water of hydration. The first mechanism is currently unpredictable, does not conform to the Hofmeister series (and Debye charge screening), and is most likely protein and salt dependent. The second two mechanisms correlate with much published work the closest of which (in the opinion of the authors) has the dual mechanisms expressed as excluded volume and preferential anion solvation by the anion.18 In this article we prefer the term competition for water than excluded volume and express the mechanism in terms of Gibbs free energy of hydration.
Conclusion
The anion results support a three mechanism model for structural stabilization of lysozyme. At low concentrations (<2 mM) the negatively charged anions bind to positively charged side chains of the protein. The result is anion specific and can either stabilize or destabilize the structure. In the case of chloride and iodide, initial addition of the anion destabilized the protein but subsequent addition stabilized it. This effect is not related to the Hofmeister series or its size (and capacity to screen charges on the protein surface) but has a measurable impact on the Tm value of the protein. At higher concentrations (>2 mM) the low charge density anions bind weakly to the protein possibly at the apolar regions of the protein.35 This reduces the Gibbs free energy associated with hydrating the newly exposed interior of the protein during unfolding. As the change in Gibbs free energy of unfolding is the sum of the energy required to break the internal bonds in the protein and hydration of the newly exposed core of the protein the reduction of the energy of hydration destabilizes the protein. As the interaction is weak this effect is concentration dependent. High charge density anions are strongly hydrated and compete for water with the lysozyme at higher concentrations (>20 mM). The competition for water increases the Gibbs free energy associated with hydrating the newly exposed interior of the protein. By increasing the Gibbs free energy of hydration kosmotropes stabilize the protein. This three mechanism model has obvious similarities to the excluded volume and preferential anion solvation explanation for protein stability.18
Materials and Methods
Hen egg white lysozyme and the salts were all sourced from Sigma Aldrich, Gillingham, UK with purities >99%. All lysozyme samples were dialyzed with HPLC grade water overnight at 4°C using Mini 8 kDa membrane dialysis kit (GE Healthcare, Amersham Place, Little Chalfont). Fresh lysozyme samples were dialyzed for each salt being tested. After dialysis the concentration of each lysozyme stock was measured using the absorbance at 280 nm on an Ultrospec 2100 pro UV spectrophotometer (Amersham Biosciences, GE Healthcare, Amersham, Buckinghamshire, UK). A 2 mg mL−1 lysozyme stock was made by appropriately diluting the lysozyme stock solution with HPLC water and adjusting to pH 7. All salts apart from NaCl were made to 2M stock solution at pH 7, the NaCl stock solution was made to 4M at pH 7. About 1 mg mL−1 lysozyme was run on the DSC for each experiment. This dilution was performed by adding the necessary amount of salt stock and HPLC water to the lysozyme stock solution to give the desired salt concentration and 1 mg mL−1 lysozyme. Nearly 1 mg mL−1 protein in HPLC water, pH 7 and varied concentrations of phosphate, sulfate, chloride, iodide, or perchlorate were used. DSC was carried out with a Nano-DSC (TA Instruments, New Castle, DE) at a heating rate of 1.5°C min−1 from 30 to 100°C in forward scans. The instrument was held at a constant temperature for 15 min between each scan. Data evaluation used the software provided by the manufacturer. Buffer–buffer baselines were subtracted from sample data. The Tm value (the temperature with the maximum heat capacity) on the DSC scan was used as a measure of protein structural stability. The precision of the DSC in determining the Tm of lysozyme in pure water was good achieving a standard deviation of only 0.02°C (n = 5) and a coefficient of variance of only 0.03%. The stability of lysozyme in various concentrations of sodium chloride was repeated see Figures 2(a) and 4 suggesting minimal run to run variation.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Hofmeister F. Zur lehre von der wirkung der slaze. Zweite mitteilung. Arch Exp Pathol Pharmakol. 1888;24:247–260. [Google Scholar]
- 2.Collins KD, Washabaugh MW. The Hofmeister effect and the behaviour of water at interfaces. Q Rev Biophys. 1985;18:323–422. doi: 10.1017/s0033583500005369. [DOI] [PubMed] [Google Scholar]
- 3.Collins KD. Charge density-dependent strength of hydration and biological structure. Biophys J. 1997;72:65–67. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Cremer PS. The inverse and direct Hofmeister series for lysozyme. Curr Opin Chem Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Cremer PS. Chemistry of Hofmeister anions and osmolytes. Annu Rev Phys Chem. 2010;61:63–83. doi: 10.1146/annurev.physchem.59.032607.093635. [DOI] [PubMed] [Google Scholar]
- 6.von Hippel PH, Wong K-Y. On the conformational stability of globular proteins. J Biol Chem. 1965;240:3909–3923. [PubMed] [Google Scholar]
- 7.Omta AW, Kropman MF, Woutersen S, Bakker HJ. Negligible effect of ions on the hydrogen bond structure in liquid water. Science. 2003;301:347–349. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 8.Omta AW, Kropman MF, Woutersen S, Bakker HJ. Influence of ions on the hydrogen-bond structure in liquid water. J Chem Phys. 2003;119:12457–12461. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 9.Batchelor JD, Olteanu A, Tripathy A, Pielak GJ. Impact of protein denaturants and stabilizers on water structure. J Am Chem Soc. 2004;126:1958–1961. doi: 10.1021/ja039335h. [DOI] [PubMed] [Google Scholar]
- 10.Bhat R, Timasheff SN. Steric exclusion is the principle source of the preferential hydration of proteins in the presence of polyethylene glycols. Protein Sci. 1992;1:1133–1143. doi: 10.1002/pro.5560010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apetri AC, Surewicz WK. Atypical effect of salts on the thermodynamic stability of human prion protein. J Biol Chem. 2003;278:22187–22192. doi: 10.1074/jbc.M302130200. [DOI] [PubMed] [Google Scholar]
- 12.Sedlak E, Stagg L, Wittung-Stafshede P. Effect of Hofmeister ions on protein thermal stability: roles of ion hydration and peptide groups? Arch Biochem Biophys. 2008;474:128–135. doi: 10.1016/j.abb.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Mazammil S, Kumar Y, Tayyab S. Anion-induced stabilization of human serum albumin prevents the formation of intermediate during urea denaturation. Proteins. 2000;40:29–38. doi: 10.1002/(sici)1097-0134(20000701)40:1<29::aid-prot50>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad F, Bigelow CC. Thermodynamic stability of protein in salt-solutions—a comparison of the effectiveness of protein stabilizers. J Prot Chem. 1986;5:355–367. [Google Scholar]
- 15.Makhatadze GI, Lopez MM, Richardson JM, Thomas ST. Anion binding to the ubiquitin molecule. Protein Sci. 1998;7:689–697. doi: 10.1002/pro.5560070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto Y, Takahashi N, Fink AL. Mechanism of acid-induced folding of proteins. Biochemistry. 1990;29:3480–3488. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- 17.Ramos CHI, Baldwin RL. Sulfate anion stabilization of native ribonuclease A both by anion binding and by the Hofmeister effect. Protein Sci. 2002;11:1771–1778. doi: 10.1110/ps.0205902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tadeo X, Pons M, Millet O. Influence of the Hofmeister anions on protein stability as studied by thermal denaturation and chemical shift perturbation. Biochemistry. 2007;46:917–923. doi: 10.1021/bi0613426. [DOI] [PubMed] [Google Scholar]
- 19.Cannon WR, Talley ND, Danzig BA, Liu XM, Martinez JS, Shreve AP, MacDonald G. Ion specific influences on the stability and unfolding transitions of a naturally aggregating protein; RecA. Biophys Chem. 2012;63:163–164. doi: 10.1016/j.bpc.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Richard AJ, Liu CC, Klinger AL, Todd MJ, Mezzasalma TM, LiCata VJ. Thermal stability landscape for Klenow DNA polymerase as a function of pH and salt concentration. Biochim Biophys Acta. 2006;1764:1546–1552. doi: 10.1016/j.bbapap.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Sikkink LA, Ramirez-Alvarado M. Salts enhance both protein stability and amyloid formation of an immunoglobulin light chain. Biophys Chem. 2008;135:25–31. doi: 10.1016/j.bpc.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komsa-Penkova R, Koynova R, Kostov G, Tenchov BJ. Thermal stability of calf skin collagen type I in salt solutions. Biochim Biophys Acta. 1996;1297:171–181. doi: 10.1016/s0167-4838(96)00092-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Cremer PS. Interactions between macromolecules and ions: the Hofmeister series. Proc Natl Acad Sci USA. 2009;106:15249–15253. doi: 10.1073/pnas.0907616106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makhatadze GI, Privalov PL. Heat-capacity of proteins. 1. Partial molar heat-capacity of individual amino-acid-residues in aqueous-solution—hydration effect. J Mol Biol. 1990;213:375–384. doi: 10.1016/S0022-2836(05)80197-4. [DOI] [PubMed] [Google Scholar]
- 25.Makhatadze GI, Privalov PL. Heat-capacity of proteins. 2. Partial molar heat-capacity of the unfolded polypeptide-chain of proteins–protein unfolding effects. J Mol Biol. 1990;213:385–391. doi: 10.1016/S0022-2836(05)80198-6. [DOI] [PubMed] [Google Scholar]
- 26.Meliga SC, Farrugia W, Ramsland PA, Falconer RJ. Cold-induced precipitation of a monoclonal IgM: a negative activation enthalpy reaction. J Phys Chem B. 2013;117:490–494. doi: 10.1021/jp309109k. [DOI] [PubMed] [Google Scholar]
- 27.Oliveberg M, Tan Y-J, Fersht AR. Negative activation enthalpies in the kinetics of protein-folding. Proc Natl Acad Sci USA. 1995;92:8926–8929. doi: 10.1073/pnas.92.19.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qvist J, Halle B. Thermal signature of hydrophobic hydration dynamics. J Am Chem Soc. 2008;130:10345–10353. doi: 10.1021/ja802668w. [DOI] [PubMed] [Google Scholar]
- 29.Frank HS, Evans MW. Free volume and entropy in condensed systems III. Entropy in binary liquid mixtures; partial molal entropy in dilute solutions; structure and thermodynamics in aqueous electrolytes. J Chem Phys. 1945;13:507–532. [Google Scholar]
- 30.Ebbinghaus S, Kim S, Heyden M, Yu X, Heugen U, Gruebele M, Leitner DM, Havenith M. An extended dynamical hydration shell around proteins. Proc Natl Acad Sci USA. 2007;104:20749–20752. doi: 10.1073/pnas.0709207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding T, Li R, Zeitler JA, Huber TL, Gladden LF, Middelberg APJ, Falconer RJ. Terahertz and far infrared spectroscopy of alanine-rich peptides having variable ellipticity. Opt Express. 2010;18:27431–27444. doi: 10.1364/OE.18.027431. [DOI] [PubMed] [Google Scholar]
- 32.Falconer RJ, Markelz AG. Terahertz spectroscopic analysis of peptides and proteins. J Infrared Millim Te. 2012;33:973–988. [Google Scholar]
- 33.Boncina M, Rescic J, Vlachy V. Solubility of lysozyme in polyethylene glycol-electrolyte mixtures: the depletion interaction and ion-specific effects. Biophys J. 2008;95:1285–1294. doi: 10.1529/biophysj.108.128694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bye JW, Cowieson NP, Cowieson AJ, Selle PH, Falconer RJ. Dual effects of sodium phytate on the structural stability and solubility of proteins. J Agric Food Chem. 2013;61:290–295. doi: 10.1021/jf303926v. [DOI] [PubMed] [Google Scholar]
- 35.Collins KD. Sticky ions in biological systems. Proc Natl Acad Sci USA. 1995;92:5553–5557. doi: 10.1073/pnas.92.12.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai Y, Winter R. Effect of molecular crowding on the temperature pressure stability diagram of ribonuclease A. ChemPhysChem. 2013;14:386–393. doi: 10.1002/cphc.201200767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


