Figure 13.
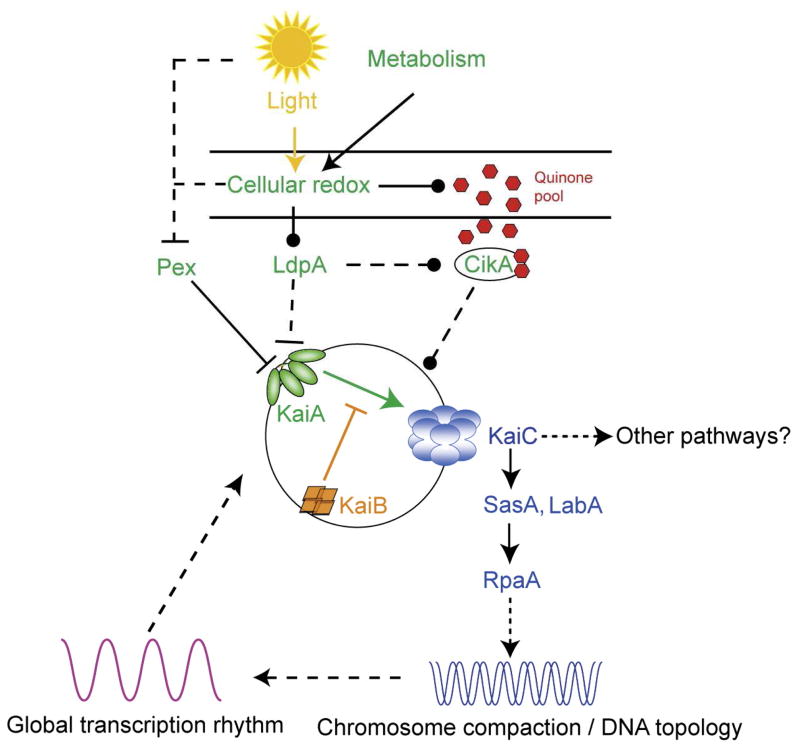
An overview of the proposed molecular mechanism of the circadian clock in S. elongatus. The central oscillator is composed of KaiA, KaiB and KaiC. KaiA stimulates KaiC phosphorylation, and KaiB inactivates KaiA when KaiC reaches a certain phosphorylation state. In the input pathway, both LdpA and CikA sense the cellular redox state, which is influenced by light and cell metabolism. LdpA affects the stabilities of CikA and KaiA through an unknown mechanism. Through its PsR domain, CikA binds quinones directly, which destabilizes CikA. CikA affects the phosphorylation states of KaiC, but where and how it works in the signal transduction pathway is not known. Pex is a transcriptional repressor of KaiA, and its abundance is sensitive to light. However, it is not clear whether the pathway that regulates pex senses light directly or does it through cellular redox. In the output pathway, SasA interacts physically with KaiC and autophosphorylates, and then transfers the phosphoryl group to RpaA, a response regulator with a DNA-binding domain. The target of RpaA has not been identified. LabA works upstream of RpaA and downstream of KaiC, but its exact function is not clear. A SasA-independent, RpaA-independent output pathway might exist. The output pathway controls DNA topology, which is proposed to regulate global gene expression. A transcription/translation rhythm could interact with and reinforce the post-translational rhythm of KaiC activities. A solid line indicates a direct effect whereas a dotted line indicates an indirect effect or an effect whose mechanism is unknown. Arrows indicate the direction of the information flow or a stimulation of activity or both. Blunt ends represent inhibition of protein activity or abundance, whereas an end with a filled circle suggests regulation of unspecified direction. Reproduced with permission for Dong and Golden, 2008.