Figure 7.
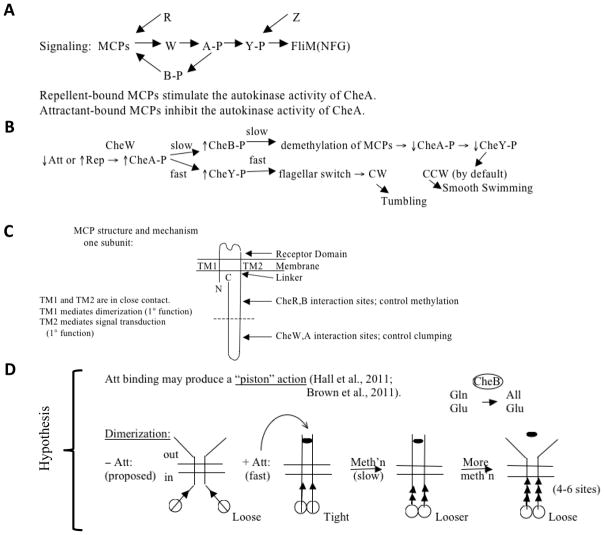
(A) and (B), Signaling pathway and mechanism of chemotactic responses to attractant (Att) and repellent (Rep) in bacteria and archaea. The model is based largely on studies with E. coli methylated chemotaxis proteins (MCPs). For explanations, see the text. CheW (W): signal transduction protein connecting the MCP signal generation with the CheA (A) sensor kinase to control its activity. CheA phosphorylates CheY (Y), and CheY phosphorylation determines the direction of flagellar rotation during activation by binding to the basal region of the flagellum, FliM(NFG). CheR (R) is the methylating S-adenosyl methionine:MCP methyltransferase while CheB (B) is the MCP methylesterase, both involved in adaptation (see also Figure 8) [see Baker and Stock, 2007; Kojadinovic et al., 2013 for reviews and comparative analyses]. CheB is phosphorylated, and thereby activated, by CheA in a slow step. CheB catalyzes deamidation, converting certain glutamines (Gln) to glutamates (Glu) so they can be methylated by CheR. Minus Att, weaker interactions are observed between intersubunit periplasmic domains and intersubunit cytoplasmic domains as compared with when Att is present. Subunit exchange occurs – Att but not + Att. Almost all amino acid replacements in TMS1 → CCW; i.e., weaker TM1:TM2 interactions, stronger intersubunit interactions, and poorer signaling. Intrasubunit movements (TM1 relative to TM2) may initiate signaling, i.e., Att binding → relative displacement of the two TMs in the membrane. CheZ is a CheY-P phosphatase. (C) Schematized view of the structure of an MCP. (D) Proposed “piston” mechanism to explain the responses of an MCP to attractant (Att➂) binding and glutamate methylation (methylated glutamates, ▲). See text for details.